How to Choose a Cell Viability or Cytotoxicity Assay
This guide describes how cell viability and cytotoxicity assays work and provides information to help you select the appropriate cell health assay for your needs. Visit the Cell Viability and Cytotoxicity Assays product listing for ordering information on the assays discussed here.
Johanna Lee and Mariel Mohns
Promega Corporation
Publication Date: 06/2019; tpub_209
Choosing a cell viability or cytotoxicity assay from among the many different options available can be a challenging task. Identifying the best cell health assay method to suit your needs requires an understanding of what each assay is measuring as a marker, how the measurement correlates with cell viability and what are the limitations of the assay chemistries. You also have the option to multiplex compatible assays to acquire more data with a statistical advantage.
Here we provide an overview of Promega cell viability and cytotoxicity assays that use plate reader-based detection methods and easy "add-mix-measure" protocols. This guide also includes key factors to consider when choosing cell health assays, so that you can sensitively compare data from well-to well, plate-to-plate, and day-to-day.
See how cell-based assays can be used to measure the number of live cells, dead cells, and cells undergoing apoptosis, autophagy or oxidative stress in cell cultures.

Introduction to Cell Viability Assays
Cell viability assays use a variety of markers as indicators of metabolically active (living) cells. Examples of markers commonly used include measuring ATP levels, measuring the ability to reduce a substrate, and detecting enzymatic/protease activities unique to living cells.
Real-time Cell Viability Assays
The RealTime-Glo™ MT Cell Viability Assay (Cat.# G9711) measures cell viability in real-time. In this assay, an engineered luciferase and a prosubstrate (which is not a substrate of luciferase) are added directly to the culture medium. The prosubstrate can penetrate cell membranes and enter cells (Figure 1). However, only viable cells with active metabolism can reduce the prosubstrate into a substrate for luciferase. The substrate then exits the cell where it is used by luciferase in the detection reagent to generate a luminescent signal. The same wells can be measured repeatedly for 3 days. The main advantages of this method are that it allows simple kinetic monitoring to determine dose response using fewer plates and cells. Also, because the method does not require cell lysis, the same cells can be used in additional cell-based assays or downstream applications.
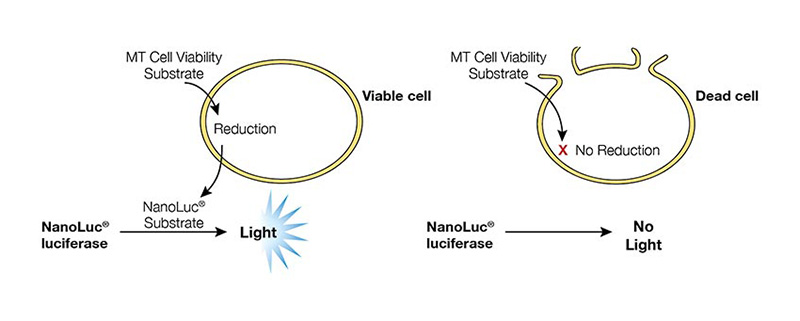
Figure 1. RealTime-Glo™ MT Cell Viability Assay overview.
ATP Cell Viability Assays
ATP can be used to measure cell viability since only viable cells can synthesize ATP. ATP can be measured using the CellTiter-Glo® Luminescent Cell Viability Assay (Cat.# G7570) with reagents containing detergent, stabilized luciferase and luciferin substrate. The detergent lyses viable cells, releasing ATP into the medium. In the presence of ATP, luciferase uses luciferin to generate luminescence, which can be detected within 10 minutes using a luminometer (Figure 2). The CellTiter-Glo® 2.0 Assay (Cat.# G9241) is provided as a single solution that reduces reagent preparation time and provides the convenience of room temperature storage for easy implementation. These ATP assays are faster than other methods since they do not require long incubation times to convert a substrate into a colored product. They also have excellent sensitivity and broad linearity, making them highly compatible with high-throughput applications where low cell numbers are used. They are also less prone to artifacts than other methods.

Figure 2. The CellTiter-Glo® Assay detects ATP as an indicator of viable cells.
Live-Cell Protease Viability Assay
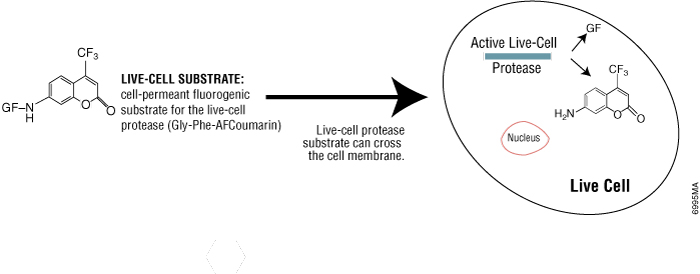
Live-cell protease activity disappears rapidly after cell death, so it is a useful marker of viable cells. Using the CellTiter-Fluor™ Cell Viability Assay (Cat.# G6080), live-cell protease activity can be measured using a cell-permeable fluorogenic protease substrate (GF-AFC). The substrate enters live cells where it is cleaved by live-cell protease to generate a fluorescent signal proportional to the number of viable cells (Figure 3). The incubation time for this method is 0.5–1 hour, which is shorter than tetrazolium assays (1–4 hours). Because this method does not lyse cells, it allows for multiplexing with many other assays in the same sample wells, including bioluminescent reporter cell-based assays.
Tetrazolium Reduction Cell Viability Assays
Tetrazolium compounds used to detect viable cells fall into two basic categories:
Positively charged compounds (MTT) that readily penetrate viable cells:
Viable cells with active metabolism are able to convert MTT into a purple-colored formazan product. Thus, color formation can be a useful marker of viable cells. The CellTiter 96® Non-Radioactive Cell Proliferation Assay (MTT) (Cat.# G4000) uses this chemistry. However, the incubation time for this method is long (usually 4 hours). Also, the formazan product is insoluble, so a solubilizing reagent must be added prior to recording absorbance readings.
Negatively charged compounds (MTS, XTT, WST-1) that do not penetrate cells:
When using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Cat.# G3582), negatively charged compounds must be combined with intermediate electron coupling reagents, which can enter cells, be reduced and then exit the cell to convert tetrazolium to the soluble formazan product. The incubation time for this method is 1–4 hours. There is no need to add a solubilizing reagent since the resulting formazan is soluble, making it more convenient.
Resazurin Reduction Cell Viability Assay
Resazurin is a cell-permeable indicator dye that is dark blue in color with little intrinsic fluorescence. The CellTiter-Blue® Cell Viability Assay (Cat.# G8080) uses resazurin to measure cell viability. Only viable cells with active metabolism can reduce resazurin into resorufin, which is pink and fluorescent. After 1–4 hours of incubation, the signal is quantified using a microplate spectrophotometer or fluorometer. This method is relatively inexpensive and more sensitive than tetrazolium assays. However, fluorescence from compounds being tested may interfere with resorufin readings.
A disadvantage of all tetrazolium or resazurin reduction assays is that they depend on the accumulation of colored or fluorescent products over time. Since the signal gradually increases over time, a decrease in cell viability during this long incubation cannot be detected.
Introduction to Cytotoxicity Assays
One of the main events that occur after cell death is the loss of membrane integrity, which allows chemicals or proteins to freely enter or exit the cell. Here we introduce cytotoxicity assays that assess the presence of dead cells by detecting outflux of certain proteins (dead-cell protease, lactate dehydrogenase) or influx of chemicals (DNA dye).
Dead-cell Protease Release Cytotoxicity Assay
When cells die and lose membrane integrity, dead-cell proteases are released. A luminogenic substrate (CytoTox-Glo™ Cytotoxicity Assay, Cat.# G9290) or fluorogenic substrate (CytoTox-Fluor™ Cytotoxicity Assay, Cat.# G9260) can then be used to measure dead-cell protease activity (Figure 4). Because the substrate is not cell-permeable, essentially no signal from this substrate is generated by intact, viable cells. Furthermore, since the assays are non-lytic, they can be multiplexed with other compatible assay chemistries.
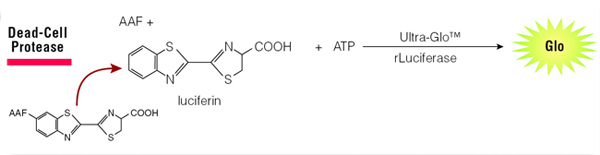
Figure 4. The CytoTox-Glo® Assay detects release of protease upon cell death.
Lactate Dehydrogenase (LDH) Release Cytotoxicity Assays
Dead cells that have lost membrane integrity release lactate dehydrogenase (LDH), which catalyzes the conversion of lactate to pyruvate with the concomitant production of NADH. Released LDH activity can be measured by providing excess substrates (lactate and NAD+) to produce NADH. This NADH can be measured using different assay chemistries:
1. LDH-Glo™ Cytotoxicity Assay
In the LDH-Glo™ Cytotoxicity Assay (Cat.# J2380), reductase uses NADH and reductase substrate (proluciferin) to generate luciferin. The luciferin is measured using a proprietary luciferase and the light signal is proportional to the amount of LDH, measured by a luminometer.

2. CytoTox-ONE™ Homogenous Membrane Integrity Assay
The CytoTox-ONE™ Homogeneous Membrane Integrity Assay (Cat.# G7890): Conversion of resazurin to a fluorescent resorufin product, measured using a fluorometer.

Figure 6. The CytoTox-ONE™ Assay detects reduction of resazurin to a fluorescent product, resorufin.
3. CytoTox 96® Non-Radioactive Cytotoxicity Assay
The CytoTox 96® Non-Radioactive Cytotoxicity Assay (Cat.# G1780) detects conversion of a tetrazolium salt (INT) into a red formazan product, measured by color absorbance

Figure 7. The CytoTox-96® Assay detects conversion of a tetrazolium salt (INT) into a red formazan product.
DNA Dye Cytotoxicity Assay
Some DNA-binding dyes are excluded from live cells, but can enter and stain the DNA of permeable dead cells. Conventional dyes, like trypan blue, often require manual counting of stained cells using a hemocytometer, which is labor-intensive and not easily scalable. Another disadvantage of conventional dyes is they may be toxic to cells and can only be used for endpoint measurement.
Newer dyes, such as the CellTox™ Green Dye, produce a fluorescent signal when bound to DNA, which is easily measured using a fluorometer. It can be diluted in culture medium and delivered directly to cells at seeding or when treating with a test compound, allowing real-time kinetic measurement. The CellTox™ Green Cytotoxicity Assay (Cat.# G8741) is nontoxic, highly photo-stable and easily scalable.
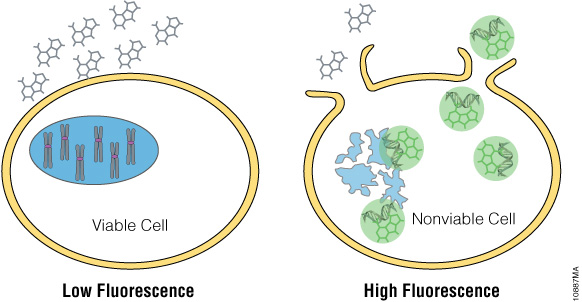
Figure 8. CellTox™ Green detects DNA released from dead cells with a fluorescent DNA binding dye.
How to Choose a Cell Health Assay
Choosing a cell health assay can be a challenging task. Here we provide a series of factors to consider when choosing cell-based assays for manual or automated systems.
What do you want to measure?
Which cells, viable or dead, do you want to detect at the end of an experiment? There are cell health assays available that specifically detect the number of living cells (viability assays), the number of dead cells (cytotoxicity assays) and for assessing the mechanism of cell death (apoptosis assays). If the information sought is simply to confirm whether there is a difference between “no treatment” negative controls and “toxin treatment” of experimental wells, the choice between measuring the number of viable cells or the number of dead cells may be irrelevant. However, if more detailed information on the mechanism of cell death is being sought, the duration of exposure to toxin, the concentration of the test compound, and the choice of the assay endpoint become critical (1). Explore the following considerations to help you determine what you want to measure and which assays can help you.
What is your model system?
The species of origin and cell types used in cell health studies are often dictated by specific project goals or the drug target that is being investigated. Regardless of the model system chosen, establishing a consistent and reproducible procedure for culturing cells and setting up assay plates is important. Variation in the number of cells per well or equilibration period prior to performing the assay may affect cellular physiology. Maintenance and handling of stock cell cultures at each step of the process should be standardized and validated for consistency. In addition, be sure to use known positive and negative controls throughout the course of the experiment to establish a general understanding of the physiological condition of the cells. Because the nature of the sample can vary depending on cell type and whether it is a 2D or 3D culture model, the assay procedure should be validated for each culture model system. A model system that requires use of smaller cell numbers, such as with primary cells or other limited cell types, will require an assay with increased sensitivity.
Are you using 2D or 3D cell cultures?
Cells in culture are only a model system and are different than cells in their normal in vivo environment. Many researchers are now using 3D cell cultures to more closely mimic in vivo conditions. Instead of growing in a monolayer on a plate surface, cells in 3D culture grow within a conformation that allows them to interact with each other, forming cell:cell connections. This added complexity can present challenges for experimental design when performing cell based assays because assay reagents may have difficulty reaching the center of large microtissues, and lytic assays may not be able to disrupt all cells within the 3D system. Assays may need to be optimized for 3D systems, by reformulating reagents with stronger detergents and incorporating mechanical disruption and longer incubation times. Colorimetric assays, such as MTT, are not optimal for use with 3D cell cultures because they have limited ability to penetrate multiple layers of cells.
The CellTiter-Glo® 3D Cell Viability Assay (Cat.# G9681) is specifically designed for determining cell viability in 3D culture models. The assay reagent has increased lytic capacity—allowing better penetration of large spheroid samples resulting in more accurate determination of viability compared to other assay methods. The CellTiter-Glo® 3D Assay Reagent measures ATP as an indicator of viability and generates a luminescent readout that is much more sensitive than colorimetric or fluorescence-based methods.
To measure cytotoxicity of a compound, the LDH-Glo™ Cytotoxicity Assay (Cat.# J2380) is ideal for use with 3D culture models. Instead of relying on penetration of the cell membrane, the assay measures LDH release from dead cells into the medium. The assay requires only small volumes of medium (2–5µl) to be removed, allowing for repeated sampling over time. This conserves microtissues and maintains remaining viable cells, allowing you to use samples for additional downstream applications, with other assays, or for nucleic acid analysis to acquire more data using the same samples.
When is the right time to perform a cell health assay?
Matching the detected assay marker to the information you need is vital to choosing the appropriate cell health assay. A basic understanding of the changes that occur during different mechanisms of cell death will help you decide which assay to choose. Figure 9 shows a simplified example illustrating chronological changes occurring during apoptosis and necrosis and the results that would be expected using assays that measure different markers.

Figure 9. Mechanisms of cell death can be determined by measuring different markers of cell viability, cytotoxicity and apoptosis in vitro.
Cells undergoing necrosis typically undergo rapid swelling, lose membrane integrity, shut down metabolism and release their cytoplasmic contents into the surrounding culture medium. Cells undergoing rapid necrosis in vitro do not have sufficient time or energy to activate apoptotic machinery and will not express apoptotic markers.
Measuring Apoptosis and Necrosis
Cultured cells that are undergoing apoptosis in vitro eventually undergo secondary necrosis. After extended incubation, apoptotic cells ultimately shut down metabolism, activate caspases, flip phosphatidylserine (PS) to the outer membrane, lose membrane integrity and release their cytoplasmic contents into the culture medium. Markers of apoptosis such as caspase activity or PS exposure on the cell surface may be present only transiently. Therefore, to determine the primary mechanism of cell death, understanding the kinetics of the cell death process in your model system is critical. To avoid missing a critical time point, you may want to choose a nonlytic real-time assay, such as the RealTime-Glo™ Annexin V Apoptosis and Necrosis Assay (Cat.# JA1011), that allows repeated readings from a single assay well over time.
Do you need to collect data from multiple time points?
Live-cell kinetic assays are detection reagents that allow the same sample well to be repeatedly measured over multiple time points. This saves you time and effort, enabling you to collect more informative data in real time. If you need to perform time and dose response experiments to determine the onset or mechanism of toxicity of a drug, you’ll likely benefit from a real-time assay. Especially if you are using limited or precious cell samples, it is critical to get as much data as possible out of your sample. Promega provides a portfolio of assays capable of live-cell kinetic cell health determination.
The RealTime-Glo™ MT Cell Viability Assay (Cat.# G9711) allows you to monitor cell viability continually in the same sample well out to 72 hours depending on cell number. The assay measures the reducing potential of viable cells and is ATP-independent, providing an orthogonal method for viability determination. The RealTime-Glo™ Annexin V Apoptosis and Necrosis Assay (Cat.# JA1011) is a plate reader-based method that measures the real-time exposure of phosphatidylserine (PS) on the outer leaflet of cell membranes during the apoptotic process. The combination and timing of luminescent (apoptotic) and fluorescent (necrotic) signals is used to differentiate secondary necrosis from necrosis caused by other cytotoxic events. The CellTox™ Green Cytotoxicity Assay (Cat.# G8741) uses a dye that produces a fluorescent signal when bound to DNA in membrane-compromised cells. This assay can be applied directly to cells at seeding or when treating with a test compound at any incubation time point, allowing real-time kinetic measurement of the onset of cytotoxicity.
Because these assays are non-toxic and non-lytic, the remaining viable cells remain intact for additional downstream applications resulting in more data per sample.
How many samples do you need to test?
If only one or a few samples need to be measured, manual counting of live or dead cells using a hemacytometer may be adequate. Imaging or flow cytometry methods are also useful for low sample numbers, but are less amenable to high-throughput applications. If large numbers of samples will be measured, homogeneous assays that do not require cell washes or centrifugation steps are the most efficient. In addition, time required for reagent preparation and incubation should be minimal. The stability of the absorbance, fluorescence or luminescence signal is another important factor that provides convenience and flexibility in recording data and minimizes variability when processing large batches of plates. For screening thousands of samples, it is beneficial to choose assays that are sensitive enough to allow miniaturization into high-density plate formats (384- or 1536-well plates).
Promega produces a broad portfolio of plate reader-based cell health assays that are fast, highly sensitive, simple and compatible with high-throughput measurement (2). While the general protocol for the majority of these "homogeneous" assays is add-mix-measure, some protocols may require transfer, incubation or agitation steps. We recommend fully reviewing the assay protocols to determine if the workflow meets your needs.
Find the Right Assay for Your Needs
Click below to compare the variety of Promega plate-based assays designed to measure cell viability, cytotoxicity and apoptosis.
What kind of sensitivity is required?
Another factor to consider when selecting an assay is sensitivity of detection. The sensitivity required is closely linked to the number of cells used per sample. In general, the luminescent assays are more sensitive than assays based on detecting fluorescence or absorbance because minimal background luminescence results in high signal-to-noise ratios. With fluorescence detection, inherent fluorescence from cells, compound fluorescence and spectral overlap between the excitation and emission wavelengths can result in reduced signal-to-noise ratios.
Detection sensitivity will vary with cell type if you choose to measure a metabolic marker, such as ATP level or MTS tetrazolium reduction. The signal-to-background ratios of some assays may be improved by increasing incubation time. The sensitivity of the detection reagent depends upon both parameters of the model system, such as the plate format and number of cells per well; in addition to the parameter being evaluated, such as a decrease in cell viability. Assays that are designed to detect a change in viability in a population of 10,000 cells may not require the most sensitive assay technology. For example, a tetrazolium assay such as the CellTiter 96® Non-Radioactive Cell Proliferation Assay (Cat.# G4000) should easily detect the difference between 10,000 and 8,000 viable cells. On the other hand, assay model systems that use low cell numbers in a high-density multiwell plate format may require maximum sensitivity of detection, such as that achieved with the CellTiter-Glo® Luminescent Cell Viability Assay (Cat.# G7570) that can easily detect fewer than 100 cells.
How stable are the reagents?
For researchers using automated high-throughput screening platforms, the reagent stability and compatibility with robotic components is often a concern. The assay reagents must be stable at ambient temperature for an adequate period of time to complete dispensing into multiple plates. In addition, the signal generated by the assay should be stable for extended periods of time to allow flexibility for recording data either consecutively or in batch mode processing. The CellTiter-Glo® 2.0 Cell Viability Assay is formulated to allow storage at ambient temperature for multiple days, and generates a stable luminescent signal with a half-life that enables batch processing.
What instruments will you need?
In some cases the choice of assay may be dictated by the availability of instrumentation to detect absorbance, fluorescence or luminescence. Our portfolio of assays contains an optional detection format for each of the three major classes of cell health assays (viability, cytotoxicity or apoptosis). In addition, results from some assays such as the CellTiter-Glo®Luminescent Cell Viability Assay (Cat.# G7570) can be recorded with more than one type of instrument (luminometer or CCD camera).
Discover the Instrument That Is Right For You
GloMax® Microplate Reader Instruments are easy-to-use, modular detection systems that offer greater flexibility when designing your plate-based assay experiments. These instruments work seamlessly with Promega assays by having preloaded protocols for easy and fast detection.
Which plates are appropriate to use?
When using our plate-based assays, choosing the right type of plate is dependent on your research needs. If microscopic observation of cells is desired, opaque clear-bottom plates are required.
In general, opaque black plates are used for reduced background for fluorescent assays, and opaque white plates are used for optimum light output for luminescent assays. However, the signal strength from most luminescent assays is adequate such that black plates can be used. Using black plates for luminescent assays provides the most flexibility for multiplexing fluorescent and luminescent assays from the same sample. Clear plates are acceptable for use with colorimetric assays, but should not be used with fluorescent or luminescent assays. Choosing the right instrument and plates can help you reduce crosstalk and other issues that might confound your data.
Do you want to perform more than one assay with your sample (multiplex)?
Multiplexing more than one assay is a versatile approach that can provide more information from a single sample. For example, you can simultaneously measure cell stress response pathway events and the mechanism of cell death in the same sample well, or measure cell viability with reporter response to normalize results to the number of live cells. Multiplexing requires compatibility of the assay chemistries being used and the ability to distinguish separate signals using different detection wavelengths or detecting different modalities such as fluorescence and luminescence. For compatible assay chemistries, multiplexing can be accomplished simultaneously in the same sample well. For some assay combinations, measurements must be done in a sequential manner. Still other multiplexing options can be enabled by separating a portion of the sample into a different container to overcome assay chemistry incompatibility, instrument requirements or plate format (e.g., clear vs. opaque plastic). For instance, the LDH-Glo™ Cytotoxicity Assay (Cat.# J2380) offers the opportunity to gather multiple data points by removing small aliquots of culture supernatant to a white opaque assay plate, thus leaving the original assay plate containing cells available for other assays such as reporter gene analysis, image analysis, nucleic acid analysis, etc.
Multiplexing assays in the sample well eliminates the need for multiple parallel samples for different assays and can save you time, effort and cost as well as provide a statistical advantage compared to using parallel samples. Several of our homogeneous cytotoxicity, apoptosis and viability assays can be multiplexed without transferring media, allowing researchers to assay multiple parameters in the same sample well.
Example multiplexing protocols:
- Interpreting Multiplexing Data Using the CellTox™ Green Cytotoxicity Assay
- An Introduction to Real Time Cell Viability Measurement
- Perform Multiplexed Cell-Based Assays on Automated Platforms
- Multiplexing Homogeneous Cell-Based Assays
Is the assay reproducible?
Reproducibility of data is an important consideration when choosing a commercial assay. However, for most cell-based assays, the variation among replicate samples is more likely to be caused by the cells rather than the assay chemistry. Variations during plating of cells can be magnified by using cell lines that tend to form clumps rather than a suspension of individual cells. Extended incubation periods and edge effects in plates may also lead to decreased reproducibility among replicates and less desirable Z’-factor values (2). When choosing an assay, understanding the causes of variability in results is key.
What is the cost?
Usually there is a trade-off between the cost of the reagent and the quality of the assay or the convenience it provides to the user. Less costly reagents often have more complex procedures, limited sensitivity, or cause toxicity to cells in culture due to longer incubations and take longer to perform. Despite those general disadvantages, there may be many applications where less costly assays are suitable; however, care should be taken to avoid reagents that are cytotoxic as they can affect the assay results or limit the ability to multiplex with other assays.
Commercially available quality-controlled reagents and assay kits are generally more expensive but often save time and cost in the long run by avoiding repeated experiments or nonreplicable results. For example, even though the reagent cost of an ATP cell viability assay may be higher than other assays, the speed (time savings), sensitivity (cell sample savings) and accuracy (confidence in data) may outweigh the initial cost. Assays with good detection sensitivity that are easier to scale down to 384- or 1536-well formats may result in savings of cell culture reagents and enable testing of very small quantities of expensive or rare test compounds and sample types, especially primary cells. Assays capable of real-time measurement can also reduce costs and save time by eliminating the need to replicate plates for time-course experiments.
Conclusion
There are many options available to help you plan and design your cell health experiments. Choosing a cell viability or cytotoxicity assay can be a challenging task, but we have a variety of solutions and resources to support your research goals. For more guidance, check out the Related Resources below. If you need more information, or need help troubleshooting your assay, our experienced Technical Services Scientists are available to help you get the results you need.
References
1. Riss, T.L. and Moravec, R.A. (2004) Use of multiple assay endpoints to investigate the effects of incubation time, dose of toxin, and plating density in cell-based cytotoxicity assays. Assay Drug Dev Technol. 2(1), 51–62.
2. Zhang, J.H., Chung, T.D. and Oldenburg, K.R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 4(2), 67–73.
MyGlo™ Reagent Reader
Speed and simplify gold-standard luminescent cell viability assays with a personal, 96-well reagent reader.