Advancing Biomedical Research with Quality Chemical Probes
The importance of high-quality chemical probes has gained increased recognition in recent years, as they play a central role in understanding complex biological mechanisms and diseases, as well as in the drug discovery process. There are numerous excellent publications discussing the various aspects of chemical probe development and use. A recent chemical probe paper highlighting the “promise & perils” of chemical probes spurred us to write this article (1). Here we highlight some key takeaways from seminal chemical probe publications and discuss the value of chemical probes, criteria for building good chemical probes, including the importance of target engagement assays, as well as resources for identifying and using high quality chemical probes.
The Importance of Chemical Probes
A chemical probe is a small molecule designed to selectively bind to and alter the function of a specific protein target (1,2). By changing a protein’s function through inhibition or stimulation, a chemical probe allows researchers to determine the protein’s role in complex living systems, such as cells and organisms. As a result, chemical probes represent a critical component of basic research and drug discovery (1,2).
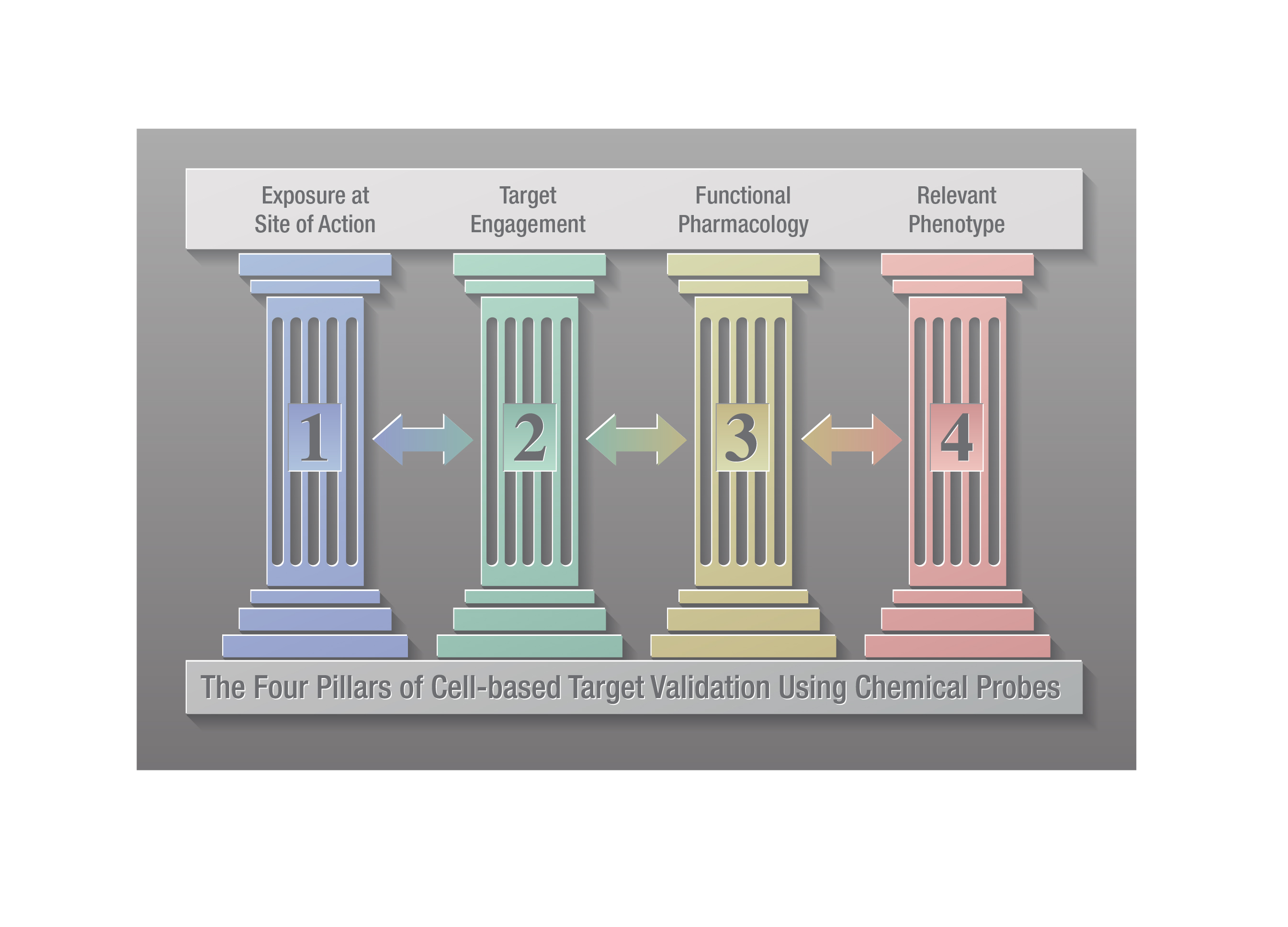
Within drug discovery, chemical probes are used alongside complementary genetic approaches such as siRNA, CRISPR and knockouts to validate the role of a target protein in disease (2,3). A four-pillared chemical probe framework has been proposed to improve cell-based target validation in drug discovery (4). The four pillars involve: 1) determining that the probe gains adequate exposure in the cell; 2) demonstrating the chemical probe engages the target (i.e., target engagement); 3) showing that the probe causes a change in target activity; and 4) demonstrating that the probe modulates relevant phenotypic changes. These authors note that measuring target engagement (pillar 2) is critical in target validation as it ties exposure (pillar 1) to functional pharmacology and phenotypic changes (pillars 3 and 4, respectively).
In addition to the central role of chemical probes in target validation, chemical probes are often used at later stages of drug discovery to help determine the clinical translatability of the target. At this later stage, probes are used in preclinical in vivo studies to aid the design of future clinical testing/monitoring. For example, chemical probes may be used to understand in vivo pharmacodynamics or identify biomarkers needed for clinical testing. This typically involves development of more specialized probes that have drug-like properties and fit the experimental need (2).
The Development of High-Quality Chemical Probes
Chemical probes need to possess specific properties to be useful for their intended purpose of addressing the role(s) of a protein target in complex biological contexts. Chemical probes must have high affinity for the intended target protein, good cellular potency, excellent selectivity over closely related target proteins and a known chemical structure and mode of inhibition. Guidance for some of these parameters is provided in the literature, such as <100nM potency in a biochemical assay, <1µM cellular potency and >30-fold selectivity over closely related proteins (3,4,5).
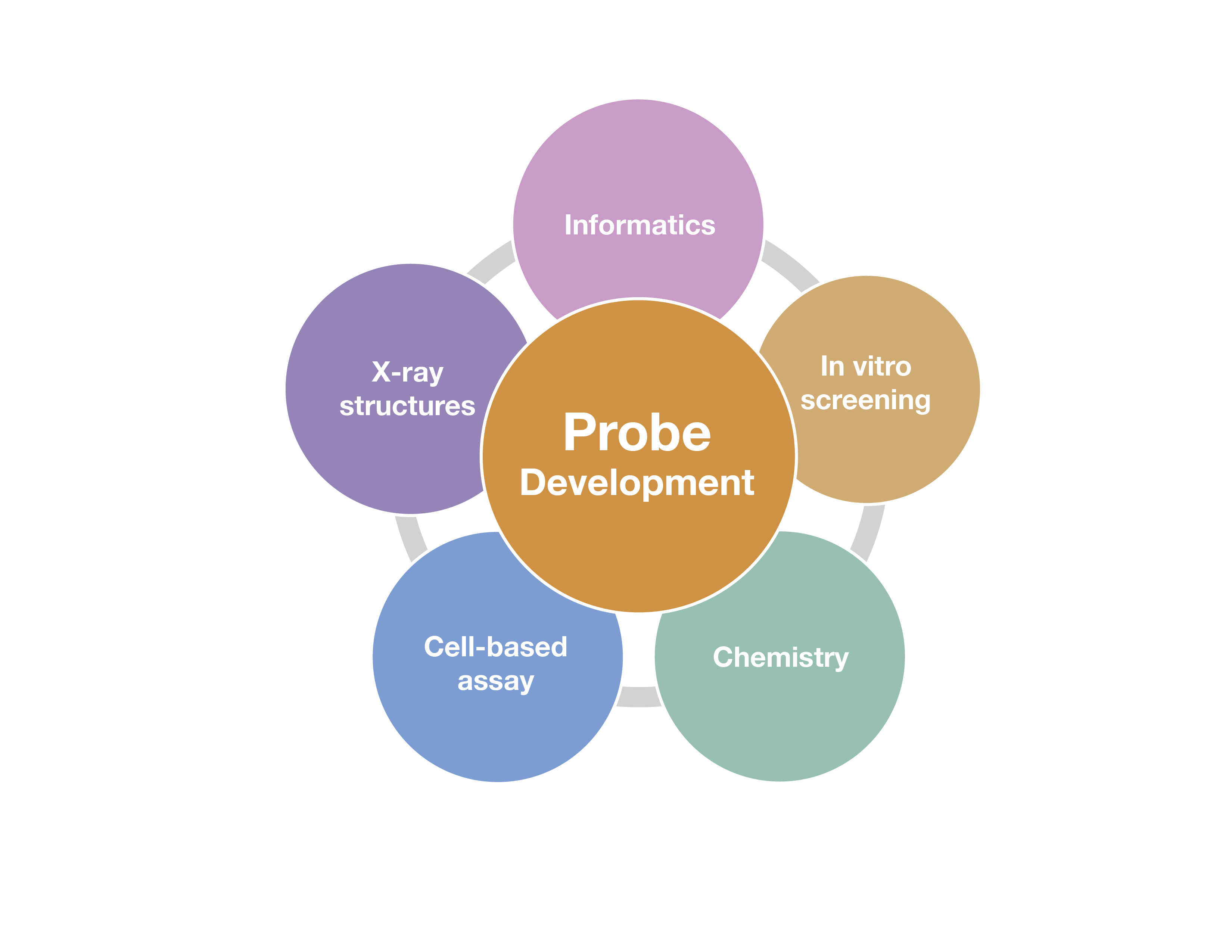
The development of high-quality chemical probes with these properties requires extensive efforts that employ multiple methods. Some of these are outlined in the figure below, and include chemistry to make and optimize the probes, screening assays to identify potential probe candidates, structural studies to evaluate the binding mode of the chemical probe candidates and cell-based assays to show the probe engages the target in cells and has an effect on a cellular phenotype.
One method utilized during chemical probe development warrants special attention. This is verifying target engagement in a cell-based assay, or the confirming that the chemical probe binds to the intended target in cells. The critical reason for measuring target engagement during chemical probe development was summarized nicely by Simon and colleagues (6):
“Without methods to confirm that chemical probes directly and selectively engage their protein targets in living systems, however, it is difficult to attribute pharmacological effects to perturbation of the protein (or proteins) of interest versus other mechanisms.”
This is important as in vitro assays are frequently performed with only the protein domain of interest, rather than the full-length protein. The authors further advocated that the most valuable target engagement assays are those that: 1) report directly on the interaction between the chemical probe and target protein, rather than a distal measurement; and 2) measure probe selectivity or the interaction of the probe with other related target proteins. Simon et al. (6) state:
“...direct measurements of target engagement should become standard practice in the development of new chemical probes…”
An Example: Chemical Probe Development for JAK3 Kinase
Among the leaders in chemical probe development are scientists at the Structural Genomics Consortium (SGC), a not-for-profit, public-private partnership formed to aid in precompetitive drug discovery. All of the chemical probe reagents and the supporting characterization data produced by the SGC are open-access and freely available to the research community. SGC chemical probes meet the high-quality standards of in vitro potency of <100nM, >30-fold selectivity relative to closely related family member proteins and demonstrate on target effects in cells <1µM (5).
One of the areas of recent research focus for the SGC is the developing kinase-specific chemical probes. Kinases are a family of >500 enzymes that play critical roles in cellular signaling, both in normal and diseased cells. As a result, kinases are key drug targets especially in cancer and inflammatory disease research. More high-quality chemical probes are needed to aid in understanding the role of multiple kinases in complex diseases. Kinases that lack potent, selective and cell-active inhibitors are referred to as the “untargeted kinome” and are the subject of research by the SGC human kinase chemical probe program (7).
Recently, members of the SGC kinase chemical probe program, in collaboration with the Laufer lab in Tuebingen, Germany, and Promega scientists, published the development of novel JAK3 kinase reversible covalent inhibitor/probe (8). The JAKs are a family of four cytoplasmic tyrosine kinases that are activated by cytokine receptor signaling and are targets for anti-inflammatory or immunosuppressive therapies. Although JAK3 is expressed primarily in hematopoetic cells, the sufficiency of a JAK3 specific inhibitor to cause effective immunosuppression is still in question. This is partly due to the ubiquitous expression of the other JAKs and to the ability to activate multiple JAK family members (such as JAK1 and JAK3) via a particular cytokine-receptor interaction.
In this Cell Chemical Biology report, the authors detail the development of a potent JAK3 inhibitor that is selective for JAK3 over other JAK family members as well as the rest of the kinome. (8). The authors achieved this by targeting a specific noncatalytic cysteine residue in the ATP binding site that is not present in other JAK family members, and only present in ten other kinases in the equivalent position. The authors first confirmed potency and selectivity in biochemical kinase activity assays. The reversible covalent nature of the binding was shown via co-crystal structures of the catalytic domain of JAK3 with the chemical probe.
Demonstrating intracellular target affinity and selectivity of the JAK3 probe was essential for its candidacy as a chemical probe. The authors used a novel BRET-based target engagement assay method (9) that assessed direct competitive binding in live cells. These assays revealed the JAK3 chemical probe to have potent apparent intracellular affinity for JAK3 (approximately 100nM) in equilibrium binding studies, and also durable but reversible intracellular binding to JAK3 in real-time cellular residence time studies. Protracted residence time profiles may be valuable for extending the in vivo pharmacokinetic effect of drug molecules (10). The functional inhibitory effect of the probe on cellular JAK3 signaling vs JAK1 was determined using cytokine-activated human T cells and monitoring phosphorylation of various STATs. This new JAK3 reversible covalent chemical probe (FM-381) will be a valuable tool in helping to elucidate the role of JAK3 over other JAK family members.
Chemical Probe Resources
Reports such as those cited here have raised awareness about the dangers of using poorly characterized inhibitors that fail to meet the high-quality guidelines for chemical probes outlined above (1,11). Unfortunately, there are inhibitors in use today that are poorly characterized or are historic compounds or both that continue to be used due to precedent. The use of low-quality inhibitors is detrimental and can lead to erroneous scientific conclusions that muddy the literature.
To assist scientists in obtaining the most accurate information about quality chemical tools available and the appropriate use of them, scientists from multiple institutes have banded together to create a website of expert curated information called the Chemical Probes Portal (1,12). The portal is designed to provide easy, free access to expert-reviewed information on chemical probes for various target proteins. The experts constitute over 100 researchers that are members of the Chemical Probes Portal’s scientific advisory board (SAB). For each chemical probe that the SAB reviews, the members provide useful information ranging from on-target and off-target activities, cellular and model organism validation, mechanism of action, recommended concentrations, where the probe can be obtained, SAB general comments and supporting references. Information on the website will grow, as chemical probe developers are invited to submit information on their probe for review and inclusion on the portal.

In addition to the Chemical Probes Portal, the SGC maintains its own website that provides extensive information on the chemical probes the SGC develops (5). Each SGC chemical probe has a multi-tab web page with relevant information about the target and the probe, including the probe’s physical and chemical properties, selectivity profile, co-crystal structures between the probe and target protein, cell-based assay data such as target engagement, recommended concentrations, references and how to obtain a sample.

The importance of chemistry and chemical probes in cancer biology has spurred the AACR (American Association for Cancer Research) to form a Chemistry in Cancer Research (CICR) working group. This group is helping to educate on the proper use of chemical probes through their web resource page and educational sessions at meetings.
Conclusions
Chemical probes are extremely valuable tools for understanding protein function in living systems in both academic and drug discovery research. However, only chemical probes that meet high performance guidelines are trustworthy and suitable for use in scientific advancement, as opposed to low quality probes, which can cause erroneous conclusions. The development of high quality chemical probes takes significant research efforts that span multiple disciplines. A central method needed during chemical probe development is target engagement, with those assays that directly measure the binding between probe and target protein in live cells being of highest value. Multiple reviews have noted that the lack of high quality chemical probes for a number of target proteins has led to spurious results. Fortunately, teams of researchers, including those involved with the SGC and Chemical Probes Portal, have taken the initiative to provide free access to detailed information and expert advice on high quality chemical probes and their use. This free guidance on the appropriate chemical probes to use for a target and their use is a valuable service to all researchers, aiding their experimental planning and their subsequent contributions to our collective understanding of the target protein.
References
- Arrowsmith, C.H. et al. (2015) The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–41.
- Garbaccio, R.M. et al. (2016) The impact of chemical probes in drug discovery: a pharmaceutical industry perspective. Cell Chem. Biol. 23, 10–17.
- Workman P. et al. (2010) Probing the probes: fitness factors for small molecule tools. Chem. Biol. 17, 561–77.
- Bunnage, M. et al. (2013) Target validation using chemical probes. Nat. Chem. Biol. 9, 195–9.
- SGC web site http://www.thesgc.org/chemical-probes
- Simon, G.M. et al. (2013) Determining target engagement in living systems. Nat. Chem. Biol. 9, 200–5.
- Fedorov, O., Miller, S. and Knapp, s. (2010) The (un)targeted cancer kinome. Nat. Chem. Biol. 29, 166–9.
- Forster, M. et al. (2016) Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket. Cell Chem. Biol. 23,1335–40.
- Robers, M. et al. (2015) Target engagement and drug residence time can be observed in living cells with BRET. Nat. Comm. 6, 10091.
- Bradshaw, J.M. et al. (2015) Prolonged and tunable residence time using reversible covalent kinase inhibitors. Nat. Chem. Biol. 11, 525–31.
- Blagg, J. et al. (2017) Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cell Cancer 32, 9–25.
- Chemical Probes Portal website www.chemicalprobes.org
Featured Application
NanoLuc® luciferase is the basis of the NanoBRET® Target Engagement assay platform that uses bioluminescence energy transfer to quantitatively measure target occupancy, compound affinity, residence time and permeability in live cells.