Sick of analyzing data? Effortlessly turn raw data into results with ProNect TPD app.
Compound Permeability and Target Affinity
Evaluating cellular permeability and affinity for target proteins and E3 ligases is fundamental to the successful development of degrader compounds. It ensures their efficacy, selectivity, safety, and mechanistic understanding, ultimately leading to more effective and targeted therapies for various diseases. Learn more about cell-based NanoBRET® Target Engagement Assays and biochemical Lumit® Assays for measuring cellular permeability and target affinity of PROTACs.
Learn about a quantitative method to assess compound cell permeability and affinity to E3 ligases using NanoBRET® Target Engagement Assays.
What Is the PROTAC's Cellular Permeability and Target Affinity?
Cell-Based Approaches
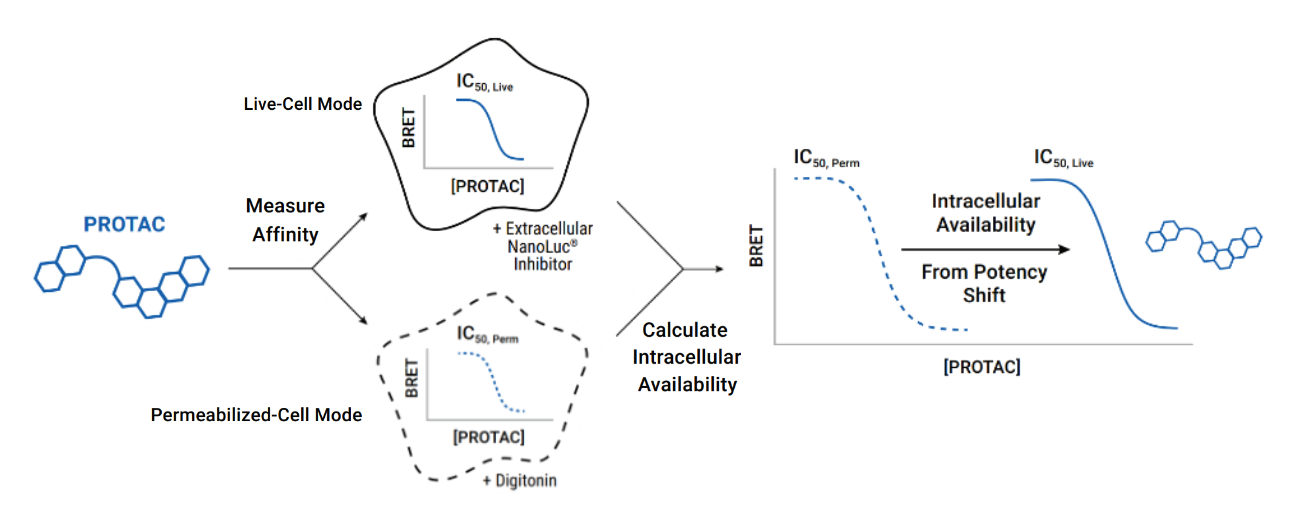
When developing degrader compounds, it is important to assess their cellular permeability and affinity for target proteins and E3 ligases. The NanoBRET® Target Engagement (TE) Assays enable the measurement of protein-small molecule binding interactions in live cells, providing quantitative determination of compound permeability, as well as target affinity and residence time.
We have successfully developed assays using NanoBRET®™ TE technology for multiple target classes important in drug discovery, including kinases, histone deacetylases (HDAC) and bromodomains. Recently, the technology has been applied to E3 ligases and NanoBRET® TE assays have been developed for CRBN, VHL, XIAP, cIAP and MDM2. The NanoBRET® TE E3 Ligase Assays can be performed in live or permeabilized cells, which allows the user to assess compound affinity for the E3 ligase as well as compound permeability.

Principle of the NanoBRET® Target Engagement Assay


Measuring cellular affinity of BET degraders targeting BRD4 using the NanoBRET® TE Assay. Comparison of cellular binding affinity for two related PROTACs that target BRD4 (left panel) using a BRD4 NanoBRET® TE Assay. Degrader affinity for the E3 ligase CRBN was compared in live and permeabilized cells using a NanoBRET® TE CRBN Assay to assess contribution of compound permeability to measured binding affinity (right panel). The study revealed that dBET6 is more permeable than dBET1 and dBET6 has slightly higher affinity for CRBN compared to dBET1.
View E3 ligase assays for CRBN and VHL or contact us for other assays (e.g., XIAP, cIAP and MDM2).
Quantifying Intracellular Availability
NanoBRET® TE technology allows determination of compound intracellular availability from the observed potency shift. Moreover, normalizing this relative shift in binding potency can be used to quantitatively assess relative intracellular availability of different compounds compared to a control.
Biochemical Approaches
We also offer biochemical methods to monitor binary interactions between small molecules and E3 ligases. In addition, these methods can measure interactions between small molecules and target proteins. The example shown here detects the interactions of molecular glues with the cereblon complex.

Schematic overview of a biochemical approach to measure interactions between small molecules and target proteins.

Molecular glue interactions with cereblon complex. To determine the relative affinities of molecular glues, we titrated them into a cereblon-thalidomide tracer complex. As the tracer is competed off, the luminescent signal decreases. Relative IC50 values correlate to target engagement assays in lysed cells.
To learn more about antibody-based approaches to study E3 ligase and protein target engagement with Lumit® technology, download our poster or contact us.
Need help with Assessing Cellular Permeability?
Learn more about our targeted protein degradation services.