Sick of analyzing data? Effortlessly turn raw data into results with ProNect TPD app.
Targeted Protein Degradation Services
Accelerate the discovery and development of degrader molecules with our comprehensive screening and profiling services.
- Comprehensive Services: Utilize cutting-edge technologies for robust data.
- Expert Collaboration: Work closely with our team to design customized experiments.
- Advanced Platforms: Leverage our experience in Targeted Protein Degradation.
- Tailored Solutions: Address unique challenges with specialized support.
- Insightful Results: Drive your project forward with reliable data and insights.
Whether you are exploring novel degraders or optimizing existing candidates, our services will deliver the results you need. Learn more about our services below or contact us for details.
Degradation Compound Profiling Services
Targeted protein degradation (TPD) is transforming drug discovery by harnessing cellular mechanisms to eliminate disease-related proteins. This approach is gaining traction as a powerful strategy for tackling historically “undruggable” targets. However, optimizing degrader compounds requires precise, quantitative insights into degradation efficiency, selectivity and cellular impact.
Our Degradation Compound Profiling Services deliver the critical data you need to understand the impact of degrader treatment on target proteins levels, whether for rapid degradation assessment of degrader efficacy or in-depth kinetic profiling. Using our advanced bioluminescent HiBiT® technology, we provide highly sensitive, end-point or real-time measurements of target protein degradation. From characterizing novel degraders to refining SAR and validating targets, our scalable, high-quality data accelerates discovery. Partner with us for reliable, reproducible and actionable insights to advance your TPD research.

Endpoint Degradation Profiling
Ideal for screening large compound libraries
- Single concentration or dose response to determine DC50 and Dmax
- Multiple time points to assess fast (5–6 hours) and sustained (18–24 hours) degradation
- Cell viability with CellTiter-Glo® assay
- Turnaround time: 3–5 weeks, depending on cell line


Kinetic Degradation Profiling
Ideal for ranking kinetic parameters of hit degraders
- Single concentration or dose response monitored for 24 hours
- Analysis of degradation rate, Dmax and Dmax50 against concentration to evaluate kinetic potency
- Endpoint cell viability with CellTiter-Glo® assay
- Turnaround time: 3–5 weeks, depending on cell line


Learn more about TPD profiling services:
Neosubstrate Panel Profiling Services
The vast majority of degraders in development and in the clinic use a small-molecule binder to the E3 ligase substrate receptor, CRBN. Upon binding, this complex can form a new protein interaction interface and induce the recruitment of a variety of neosubstrates, leading to their ubiquitination and degradation.
While most CRBN binders are derivatives of thalidomide, the chemical and structural determinants of neosubstrate selectivity remain a topic of intense study. CRBN-recruiting molecular glues or heterobifunctional PROTACs that lack specificity for their intended target and induce significant off-target neosubstrate degradation pose safety concerns in clinical applications.
The six most commonly degraded neosubstrates (GSPT1, CK1a, IKZF1, IKZF2, IKZF3 and SALL4) are associated with off-target toxicity, hematological toxicities such as neutropenia and thrombocytopenia, and teratogenicity. Therefore, early screening for degradation of these six neosubstrates in clinical degrader programs can aid optimization to enhance selectivity and de-risk compounds moving forward into the clinic.
HiBiT is an 11-amino-acid peptide tag that binds to its complementation partner, LgBiT, forming the functional NanoBiT® luciferase, which produces a bright bioluminescent signal with over 7 logs of linear dynamic range. For neosubstrate degradation studies, HiBiT is genomically integrated at endogenous loci, enabling sensitive, quantitative detection of protein levels via luminescence. Degradation is assessed 24 hours after treatment with a degrader compound, and the DC50, indicating the half-maximal response, is calculated.


Detecting targeted protein degradation using HiBiT tagging. The target HiBiT fusion protein interacts with LgBiT (either provided in solution or expressed in cells) to generate a luminescent signal. In the presence of a degrader compound, the luminescent signal decreases as the target is degraded over time. A similar degradation profile can be obtained using NanoLuc® fusions.
Neosubstrate Panel for CRBN-Based Degrader Selectivity Profiling






Neosubstrate panel for molecular glue degradation selectivity. Six neosubstrate targets for IMiD molecular glues were established as HiBiT CRISPR knock-in cell lines across different cell types: IKZF1 and IKZF2 (Jurkat), IKZF3 (MM.1S), SALL 4 (SK-N-DZ), CK1α and GSPT1 (HEK293). This neosubstrate panel enables screening for molecular glue library degradation selectivity and off-target effects with CRBN-recruiting PROTACs. Recommended endpoints for screening include 6 and 24 hours, with concurrent cell viability counter-screen. RLU values were normalized using the CellTiter-Glo® Luminescent Cell Viability Assay.
|
Compound |
Primary Neosubstrate(s) |
Indication |
Off-Target Risks |
|---|---|---|---|
|
DKY-709 |
IKZF2 (Helios) |
Solid tumors (immunotherapy) |
Potential off-target degradation of SALL4 and IKZF4 |
|
SJ6986 |
GSPT1, GSPT2 |
Acute leukemia |
Potential off-target degradation of SALL4 or other CRBN substrates, leading to hematological toxicities (e.g., neutropenia, thrombocytopenia) |
|
Thalidomide |
Broad, including IKZF1, IKZF3 |
Multiple myeloma, leprosy, inflammatory diseases |
Teratogenicity (SALL4 degradation), peripheral neuropathy, and hematological toxicities |
|
DEG-77 |
IKZF2 (Helios), CK1α |
Acute myeloid leukemia (preclinical) |
Risk of hematological toxicities (e.g., neutropenia, thrombocytopenia) and possible off-target effects on other CRBN neosubstrates |
|
Iberdomide |
IKZF1 (Ikaros), IKZF3 (Aiolos) |
Multiple myeloma, autoimmune diseases |
Improved specificity reduces risks, but teratogenicity (SALL4 degradation) and hematological toxicity remain concerns |
|
Lenalidomide |
IKZF1, IKZF3 |
Multiple myeloma, myelodysplastic syndrome |
Teratogenicity (SALL4 degradation), hematological toxicities (neutropenia, thrombocytopenia), and thromboembolism risks |
Description of Services
Assay Format
- Compounds profiled in dose-response curves, using 10-point, 3-fold dilution series
Data you receive
- Dose-response curves for your compounds
- Calculated DC50 for your compounds
- Known degrader control compound dose-response curve and DC50 value
Assay Timepoint Options
- Viability measurements with CellTiter-Glo® 2.0 at 6 and 24 hours
- Viability measurements with CellTiter-Glo® 2.0 at 24 hours
Service Schedule
- Service offered on a monthly basis
- Turnaround time: 2–3 weeks
Permeability and Affinity Services
Successful targeted protein degradation (TPD) requires not only inducing protein loss but also efficiently reaching and binding targets within cells. Permeability determines how well a degrader crosses membranes to access intracellular targets, while affinity dictates the strength and specificity of interactions with key proteins like E3 ligases and target proteins. Optimizing these properties enhances degrader potency, selectivity, and therapeutic efficacy and is especially critical for non-conventional degraders beyond the rule of five (Ro5), such as PROTACs, which defy traditional small molecule standards.
Our Permeability and Affinity Services provide the quantitative insights needed to refine degrader design and improve drug-like properties. Using advanced cell-based assays, we assess permeability to predict intracellular availability and measure binding affinity for efficient target engagement. Whether optimizing scaffold design, assessing SAR or comparing candidates, our high-quality, reproducible data accelerate TPD research. Partner with us for precise, actionable insights to streamline degrader development.
NanoBRET® Target Engagement and Cellular Permeability
- De novo assay and tracer development
- Assessment of degrader binding affinity to target and E3 (e.g., CRBN, VHL, XIAP, cIAP and MDM2)
- Assessment of degrader permeability in live vs. permeabilized cells
- Turnaround time (profiling): 2–3 weeks


Degrader Results with Automated VHL TE Assay
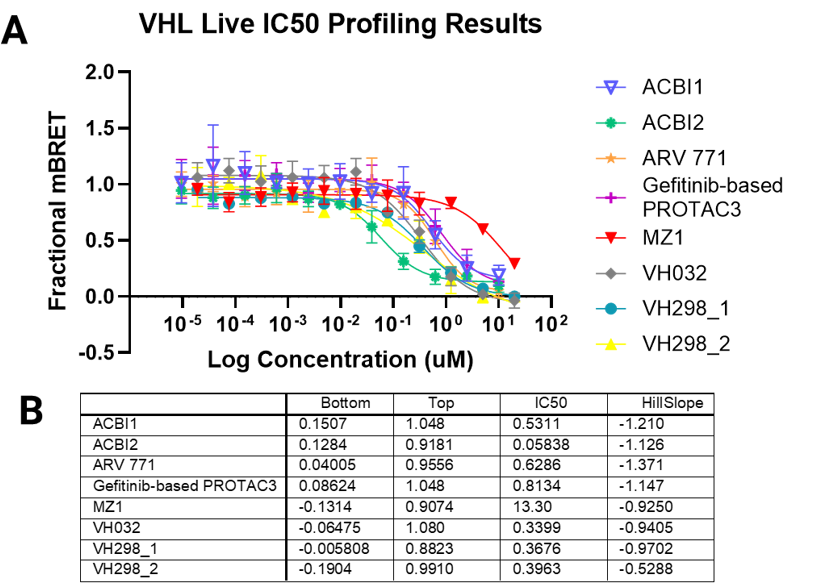
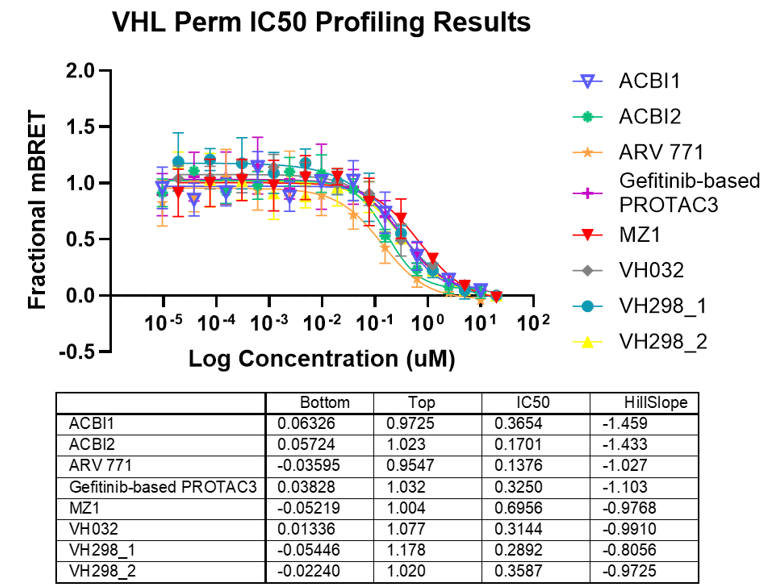
Panel A. Compound concentration-response curves from NanoBRET® TE VHL assays in live-cell or permeabilized-cell modes, respectively. VH298 was used as a permeable control compound. Panel B. Lower and upper asymptote, IC50, and Hill slope parameters calculated from non-linear regression curves.
VHL TE Assay Permeability Parameters
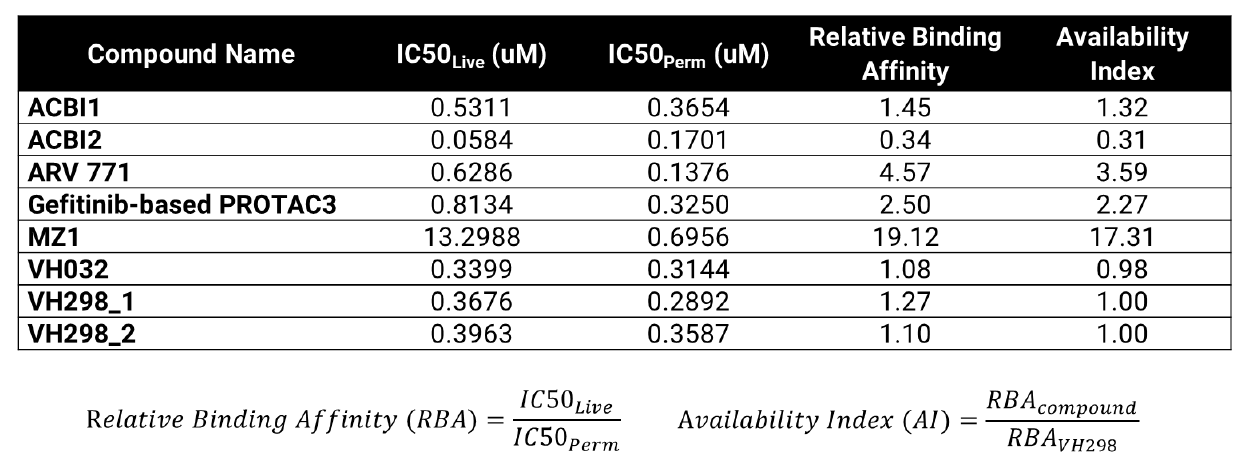
Calculated relative binding affinity (RBA) and availability index (AI) values for each compound. An RBA value of approximately 1 indicates similar free concentrations inside and outside of the cell. AI >1 suggests reduced availability inside the cell compared to VH298 control. AI <1 could indicate compound accumulates in cells at higher relative concentrations compared to VH298 control.
Ternary Complex Formation Services
Ternary complex formation—involving the degrader, target protein and E3 ligase—is a key step in driving protein elimination. The stability and dynamics of this complex directly impact degrader potency, selectivity and cellular efficacy. Understanding how efficiently a degrader promotes ternary complex formation and how long this interaction persists is essential for optimizing degrader function and advancing lead compounds.
Our Ternary Complex Formation Services provide quantitative, real-time insights into degrader-induced interactions using our highly sensitive NanoBRET® technology. Our assays precisely measure complex formation in live cells helping researchers refine degrader design. Whether prioritizing compounds, optimizing SAR or investigating mechanism of action, our services deliver reliable, high-quality data to accelerate TPD research. Partner with us for actionable insights that drive therapeutic innovation.
NanoBRET® Ternary Complex Formation
- De novo assay development and testing
- Degrader profiling in endpoint or kinetic format
- Single concentration or dose response
- Turnaround time (profiling): 3–4 weeks

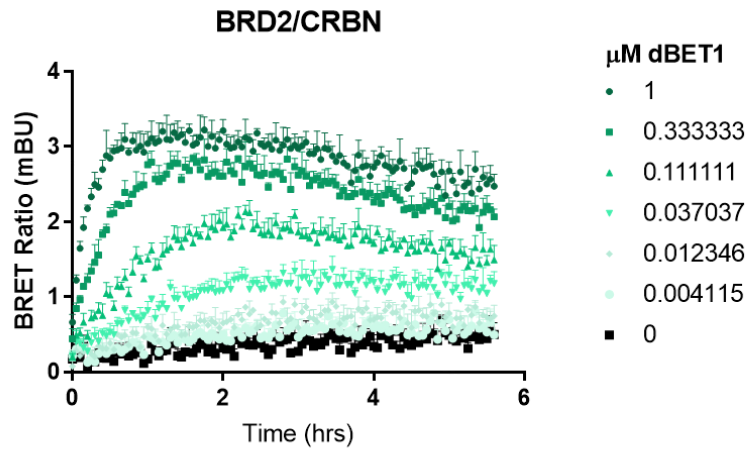
Screening for Neosubstrate Ternary Complex Formation with Transient NanoBRET® Assay


NanoBRET® Ternary Complex Assay optimization for common neosubstrates and CRBN. NanoLuc® neosubstrate and HaloTag® CRBN fusions were expressed in HEK293 cells with or without protease inhibitor MG132. All N- and C- orientations were tested at 1:10 and 1:100 ratios. Results show a significant increase in BRET upon addition of molecular glue due to formation of ternary complex.
Ubiquitination Services
Ubiquitination of the target protein is a crucial step for recognition and degradation by the proteasome. Inefficient or incomplete ubiquitination can limit degrader efficacy, making it essential to characterize its extent, kinetics and selectivity. Understanding this process provides key insights into degrader mechanism of action and helps optimize compound design for potency and selectivity.
Our Ubiquitination Services use advanced NanoBRET® technology to deliver real-time, quantitative measurements of target ubiquitination in live cells. Our assays precisely assess ubiquitination efficiency, helping researchers evaluate degrader performance, optimize SAR and refine compound selection. Whether comparing degrader candidates, investigating E3 ligase specificity or fine-tuning potency, our services provide high-quality, reproducible data to accelerate TPD research. Partner with us for critical insights into ubiquitination dynamics and next-generation degrader development.
NanoBRET® Target Ubiquitination
- De novo assay development and testing
- Degrader profiling in endpoint or kinetic format
- Single concentration or dose response
- Turnaround time (profiling): 3–4 weeks

