Cell Line Authentication Guide
Your cell lines could be at risk
Human cultured cell lines are used in many biomedical research and clinical applications, including cancer research, drug discovery, genetics and biobanking. However, misidentification of cell lines continues to plague research, despite prominent scientists calling for authentication. Today, many journals require cell line authentication for publication.

Why should I care about misidentified cell lines?
- Misidentified cell lines could lead to invalidation of data, as well as lost time, money and effort. In fact, studies have estimated that up to 20% of published papers could be invalid due to misidentified or cross-contaminated cell lines.
- Since 2016, many journals and funding agencies have started requiring authentication of human cell lines prior to paper or grant submission.
- It’s just bad science!

When should I authenticate a cell line?
- When establishing or acquiring a new cell line
- Within the first week of passaging a new cell culture
- When starting a new series of experiments
- When routinely passaging cell lines
- When observing inconsistent cell behavior or unexpected results
- When preparing to publish
- When freezing cell stocks to verify purity
How does Cell Line Authentication work?
Cell Line Authentication relies on the analysis of Short Tandem Repeats (STRs). STRs are repetitive sequence elements 2-7 base pairs long that are located throughout the human genome. These sequences are polymorphic – the specific pattern of a locus can be repeated any number of times.
In 2011, a committee of experts published the ANSI/ATCC ASN-0002-2011 consensus guidelines for best practices in cell line authentication using STR genotyping. First, STRs are amplified by PCR using primers outside the repeat sequence. The resulting amplicons are separated using capillary electrophoresis to determine the number of repeats in the sample. The resulting profile is then compared with a reference sample to verify the origin of the cell line. The statistical power of discrimination increases as you amplify more loci, due to the repeat variation among humans and human cell lines derived from individuals.
The updated ASN-0002 Revised 2022 recommends the following set of 13 autosomal STR loci: CSF1PO, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, FGA, TH01, TPOX and vWA.

What are the steps in Cell Line Authentication?
- Check the cell line name against the database of misidentified cell lines.
- Perform STR profile testing.
- Compare STR profile to donor or database
- Determine the percent match.
Verify that any cell lines intended to use are not listed in the ICLAC Register of Misidentified Cell Lines. Checking this database is an easy and free way to avoid loss of research time and funds spent pursuing experiments with erroneously labeled cell lines.
Perform STR genotyping on DNA purified from the cell line of interest, or from cells conserved on sample storage cards. Alternatively, samples can be sent to a high-quality core facility that follows the ANSI/ATCC guidelines.
Compare the obtained genotype to a reference database, like the ATCC STR Database, the DSMZ STR Profile Database, or Cellosaurus. Each of these databases includes a search option to compare your obtained test sample genotype to the cell lines included in the database. Cell lines are typically aneuploid and may show genetic drift relative to the reference. While this should be minimized with good cell culture practices, some variability is expected. Therefore, a threshold of 80% genotype match has been established to claim cell line authentication.
Examine any cell lines for cross-contamination based on the indicated genotype. Depending on the Match Formulation used, extra alleles present due to cross-contamination would lower the percent match. In contrast, extra alleles in the test sample do not affect the percent match calculated. The ANSI/ATCC Guidelines indicate that it is unlikely that any single cell line would have more than two alleles in three or more loci. Multiple alleles at multiple loci are another indication of cross-contamination.
Testing alone cannot substitute for good cell culture practices, including a written lab policy and training protocol. In addition to performing STR genotyping for cell line authentication, consider additional measures to avoid other types of contamination, such as regular mycoplasma testing and testing for mouse cell contamination if mouse cell lines are used in the lab. Several companies and core facilities offer reagents and fee-for-service mycoplasma testing.

How can I authenticate my cell lines in my lab?
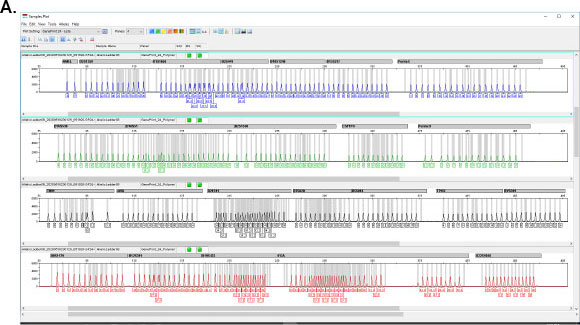
GenePrint® 24 System
The GenePrint® 24 System provides a convenient STR-based method for detecting cell and tissue mixtures and confirming cell origin and identity. Other applications include twin zygosity testing, tissue and cell provenance testing, floater analysis, and analysis of genetic changes during cancer development and progression.
- Maximum power of discrimination
- Resolves complex mixtures from related individuals or multiple sources
- Optimal heterozygote balance using up to 5ng of DNA
- Better consistency in detecting low-level mixtures
- Amplifies all recommend ANSI-0002, Revised 2021 loci
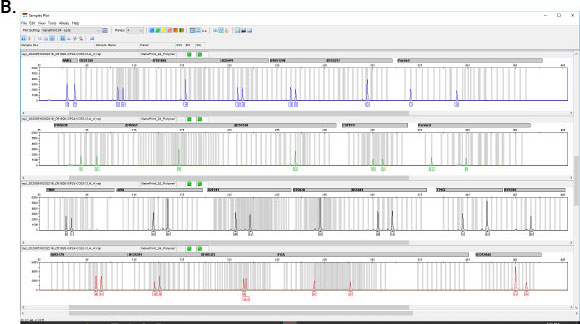
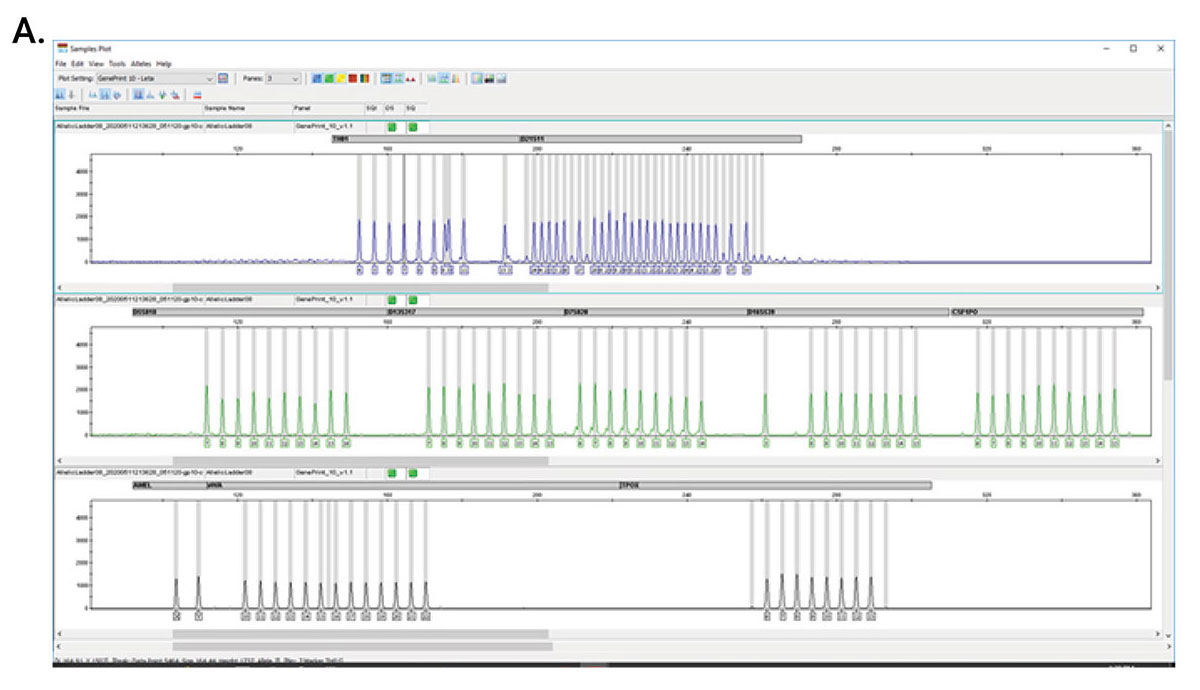
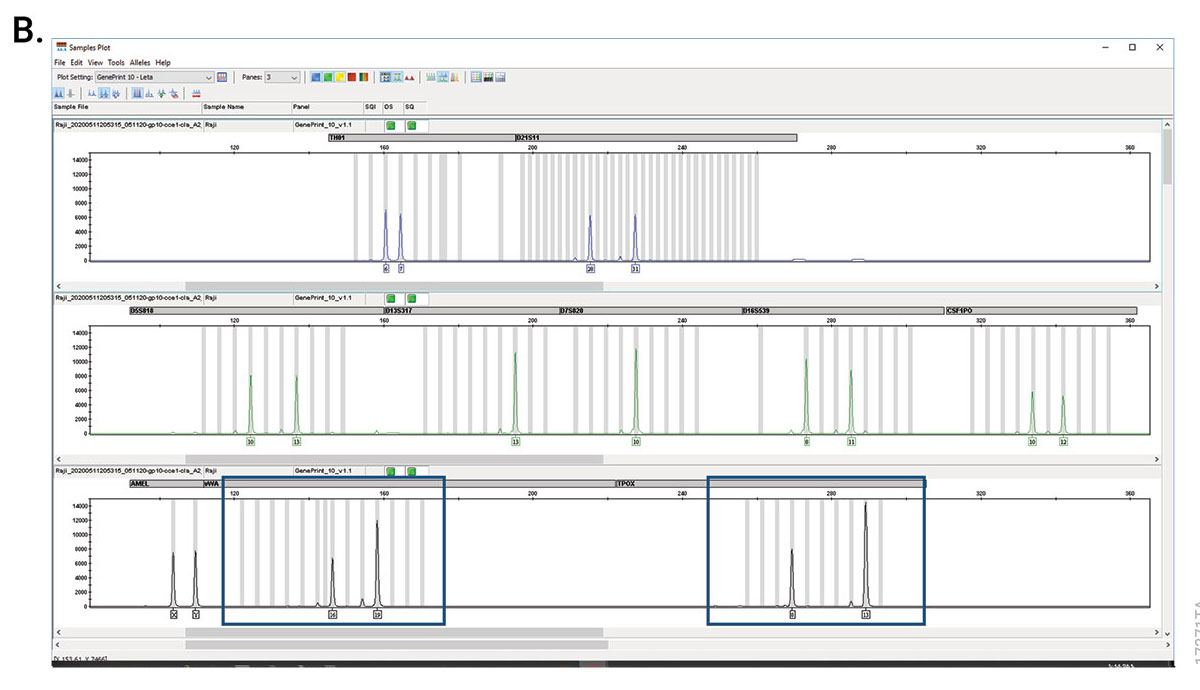
GenePrint® 10 System
The GenePrint® 10 System is a 10-locus multiplex system designed to generate a multi-locus DNA profile by amplifying human genomic DNA extracted from cell pellets or cell culture enrichment media, or by direct amplification from storage cards.
- Tolerates higher DNA amounts
- Better balance for aneuploid samples
- Amplification in less than 1.5 hours
- Lower number of loci simplifies interpretation

Spectrum Compact CE System
The Spectrum Compact CE System is a benchtop capillary electrophoresis instrument that supports cell line authentication using common STR systems, including the GenePrint® 24 system and GenePrint® 10 system. The instrument offers flexible run scheduling and an easy-to-use interface, so you’ll never have to send your samples for external processing again.
Additional Resources
Webinar: Maintaining Scientific Integrity Through Cell Line Authentication
This webinar provides a brief history of cell line contamination and outlines a simple and inexpensive method for authenticating human cell lines.
Application Note
Cell Line Authentication Using the GenePrint® 10 and GenePrint® 24 Systems This application note includes an overview of the process for cell line authentication, describes best practices for determining cell line identity, and presents methods and results for using the GenePrint® Systems for STR amplification and the Spectrum Compact CE System for analysis.
List of Journals requiring Cell Line Authentication
The publications below require some level of cell line authentication before accepting research articles. Note: This list may not be complete.
- Cancer Discovery
- Cancer Research
- Clinical Cancer Research
- Cancer Epidemiology, Biomarkers & Prevention
- Molecular Cancer Research
- Molecular Cancer Therapeutics
- Cancer Prevention Research
- Endocrinology
- Endocrine Reviews
- Journal of Clinical Endocrinology & Metabolism
- Molecular Endocrinology
- Hormones and Cancer
- Journal of Endocrinology
- Journal of Molecular Endocrinology
- Endocrine-Related Cancer
- Nature Reviews Molecular Cell Biology
- Nature
- Nature Genetics
- Nature Reviews Immunology
- Nature Reviews Cancer
- Nature Reviews Neuroscience
- Nature Biotechnology
- Nature Methods
Where can I find an STR Profile Testing Service?
These laboratories offer various levels of cell line authentication services using Promega STR systems.
