Direct Amplification of Blood Deposited on Substrates Commonly Encountered at Crime Scenes Using the PowerPlex® 18D and PowerPlex® Fusion Systems
The Pennsylvania State University, Forensic Science Program
Publication Date: 2016

Introduction
Autosomal STR testing is routinely performed in forensic crime laboratories (1) (2) (3) . Evidence such as clothing items, drinking straws, cigarette butts, leaves or wood commonly encountered at crime scenes often contain body fluids. Direct amplification of blood and saliva deposited on FTA® and nonFTA paper substrates can successfully yield profiles with direct and nondirect amplification kits (4) (5) (6) (7) . Direct amplification can be performed using bodily fluids deposited on various types of swabs that do not contain lysing reagents after the swabs are pretreated with washing reagents (5) .
The goal of this research was to generate autosomal short tandem repeat (STR) profiles from bloodstains found on substrates that are typically collected from crime scenes and submitted for analysis in criminalistics laboratories. This study included single-source blood samples. PowerPlex® 18D and PowerPlex® Fusion Systems (Promega Corporation, Madison, WI, USA) were chosen for this study as these kits are designed for direct amplification of autosomal loci. This study was expanded upon to include amplification from saliva samples in a 2016 paper (8) .
Materials and Methods
Substrates used to simulate crime scene evidence: As illustrated in Figure 1, the ten substrates used in this research included five different types of fabric ranging from 100% white cotton and blue denim jeans (100% cotton) to cotton/polyester blend materials of various colors. Five other objects were chosen as they are often encountered in indoor and outdoor crime scenes: black leather, white drinking straw, dry brown leaf, cigarette butt and a piece of a wood chip. None of these objects contained any lysing agent. Some of these objects may also contain potential inhibitors. (7) All items, except the leaf and the wood chip, which were picked up from the grounds around campus, were obtained from commercial sources. None of the substrates were sterilized or cleaned in any way.
Sample collection from humans: All samples were collected following the guidelines as dictated by the policy of the Institutional Review Board (IRB) and Institutional Biosafety Committee (IBC) at The Pennsylvania State University. Blood samples from two deceased males were collected in sterile tubes containing EDTA, an anticoagulation agent to prevent DNA degradation, and kept frozen for three years until ready for deposition. Both donors were anonymized and samples were designated as M1 and M2.
Autosomal STR Typing Protocols
DNA extraction and quantitation of reference samples: Liquid blood was thoroughly mixed before depositing 0.1µl of each sample on the ten substrates shown in Figure 1. All items were left to dry at room temperature for 24 hours prior to processing. Each was then extracted using the prepGEM® DNA Extraction Kit (ZyGEM NZ Ltd, Hamilton, NZ) following the manufacturer’s protocol (9) . All extracted DNA was quantified with the Quantifiler® Human DNA Quantification Kit (Life Technologies, Foster City, CA, USA) and the Applied Biosystems 7500 Real Time PCR System (Applied Biosystems, Foster City, California, USA) following the recommended protocol (10) . The concentration of DNA in these samples ranged from 0.01ng/µl to 0.73ng/µl.
 Figure 1. The substrate types used in these experiments.
Figure 1. The substrate types used in these experiments. Blood from M1 and M2 samples were deposited on each of the ten substrates depicted in Figure 1. First row from left to right were the following fabric samples: 100% white cotton, green cotton/polyester blend, gray cotton/polyester blend, red cotton/polyester blend and 100% cotton denim jeans. Second row from left to right: black leather, white drinking straw, brown leaf, wood chip and cigarette butt. Third row: Harris Micro-Punch along with a bloodstained fabric and a glass slide on which stains were placed for punching. Slides were changed between substrates. Leaves, cigarette butt and straws were punched on a Harris punch mat and did not require a glass slide. Only 0.1µl of blood was deposited on each substrate. Each substrate containing the bloodstain was then placed on a glass slide. Each stain was punched to create a 1.2mm punch using the Harris 1.2mm puncher. The puncher was placed on the stain and rotated clockwise and anticlockwise several times until a punch was generated. Each created punch containing bloodstain was then put into a sterile 0.2ml tube, and amplification reagents were added to each tube.
STR typing protocols for simulated crime scene samples: Using sterile aerosol pipette tips, 0.1µl of blood from the two donors was deposited on all ten substrates and left at room temperature to dry for 24 hours. A Harris Micro-Punch 1.2mm punching device was used to create a 1.2mm punch of each bloodstain as shown in Figure 1. All of these bloodstains were punched using the Micro-Punch device except the wood chip. Since it was difficult to punch the wood chip, a minute piece, approximating 1.2mm in size, was shaved off from this bloodstain. A preliminary study was conducted using bloodstains from M1 sample deposited on white cotton, green, gray and red polyester/cotton blend fabric and denim jeans. Each of these stains was pretreated with either the PunchSolution™ or SwabSolution™ Reagent. In all subsequent experiments, none of the bloodstained punches were pretreated with reagents or buffer.
Amplification kits: An optimal amount of DNA from each extracted reference sample was amplified with the two direct autosomal STR amplification kits: PowerPlex® 18D and PowerPlex® Fusion Systems. Manufacturer’s recommendations were followed except that the extracted DNA was amplified in a reduced reaction volume of 12.5µl. Amplification parameters were the same as those recommended by the kit manufacturer (11) (12) . At first, attempts were made to amplify the bloodstains in a 12.5µl reaction volume. Larger loci dropped out more often, noise level was high and the quality of the profiles was not as good when the reaction volume was less than the recommended volumes. Thus the simulated crime scene samples were amplified directly in a 25µl reaction volume. Each of the punched substrates and the shaved wood chip remained in the amplification reaction mixture during the thermal cycling process. Each DNA extract and the punched substrates containing the bloodstains were amplified in duplicate. When necessary, some of these samples were amplified multiple times. Thermal cycling conditions were the same as those recommended by the manufacturer.
Data analysis: The amplified DNA fragments were analyzed using capillary electrophoresis on the Applied Biosystems® 3130xl Genetic Analyzer. Analyses of the generated STR profiles were performed using GeneMarker®HID Software, Version 2.7.1, from SoftGenetics LLC (State College, PA, USA).
Results and Discussion
The PowerPlex® 18D and PowerPlex® Fusion profiles were consistent with the reference profile of the donor amplified with the same kit. Representative results are shown in Figure 2.
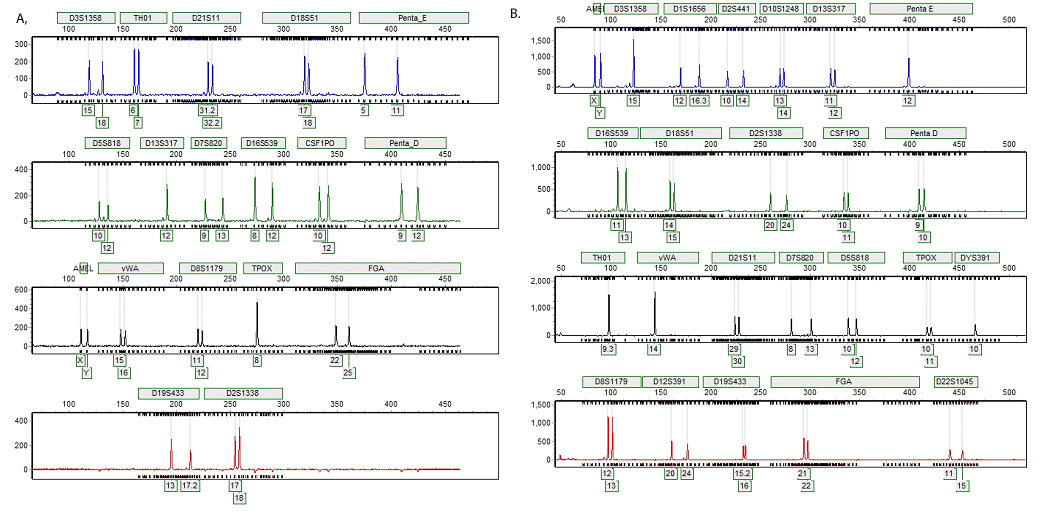 Figure 2. Representative STR profiles generated from direct amplification of DNA from 0.1µl of blood.
Figure 2. Representative STR profiles generated from direct amplification of DNA from 0.1µl of blood. Panel A. Bloodstain on blue denim jeans created with 0.1µl of blood from donor M1 and punched with the Harris 1.2mm Micro-Punch yielded a complete STR profile when amplified with the PowerPlex® 18D System. The substrate remained in the amplification reaction during the thermal cycling process. Panel B. Bloodstain on a white straw created with 0.1µl of blood from donor M2 and punched with the Harris 1.2mm Micro-Punch yielded a complete STR profile when amplified with the PowerPlex® Fusion Amplification System. The substrate remained in the amplification reaction during the thermal cycling process.
A preliminary study was conducted using blood from M1 donor deposited on five fabric samples mentioned above. Each stain was pretreated with either the PunchSolution™ or SwabSolution™ Reagent. The results of this study were compared to the results obtained from bloodstains deposited on the same fabric pieces and amplified directly without any pretreatment. The quality of the peak morphology and peak height ratios for the profiles was found to be similar and dropout of alleles occurred similarly whether the samples were pretreated or not. Thus, in all subsequent experiments, none of the bloodstained punches were pretreated with either the PunchSolution™ or SwabSolution™ Reagent.
Complete and concordant autosomal STR profiles were successfully obtained from the bloodstains deposited on these ten challenging objects when the body fluid was amplified directly using the PowerPlex® 18D and PowerPlex® Fusion Systems. The profiles were consistent within and between the substrates. DNA extracted from the bloodstains deposited on the same substrates using traditional method of extraction was amplified with both kits. These 20 profiles generated from the extracted DNA were consistent with the profiles obtained when bloodstains on the substrates were amplified directly. The quality of the profiles was also similar (data not shown).
The ten substrates chosen for this study are commonly encountered at crime scenes and often submitted to forensic science laboratories for analysis. This research was undertaken to determine if the primers contained in the two PowerPlex® Systems can generate autosomal profiles by direct amplification when the substrates containing bloodstains were punched or shaved and remained in the amplification reagents during the thermal cycling process. As indicated above, results were similar when treated and untreated stains were amplified with these two kits. None of substrates contained any lysing reagent, and some of the substrates contained potential inhibitors.
As shown in Figure 3, generation of complete profiles was not successful for every attempted amplification. This was particularly true when the bloodstains deposited on cigarette butts and wood chips were amplified. With the PowerPlex® 18D System, full profiles were generated from duplicate bloodstains deposited on green fabric, white gauze, black leather, blue denim jean and a brown leaf. Bloodstains on wood chips and drinking straws did not always yield complete profiles. However, either partial (allele dropout at 1+ loci) or full profiles were obtained when multiple amplifications of these samples were attempted. Although full profiles were obtained from bloodstains on red fabric, grey fabric and cigarette butts, these items occasionally yielded no results when the bloodstains on these objects were amplified multiple times. With the PowerPlex® Fusion System, full profiles were generated from duplicate bloodstains deposited on gray fabric, white gauze, blue denim jean and a drinking straw. Bloodstains on red fabric, green fabric, black leather and a cigarette butt did not always yield complete profiles. However, either partial or full profiles were obtained when multiple amplifications of these items were attempted. Although, full profiles were obtained from bloodstains on brown leaf and wood chip, these items occasionally yielded no results when amplified multiple times. Similar results were obtained with extracted DNA (data not shown). Failure to amplify directly could be due to possible inhibitory factors found in some of the substrates used in this study.
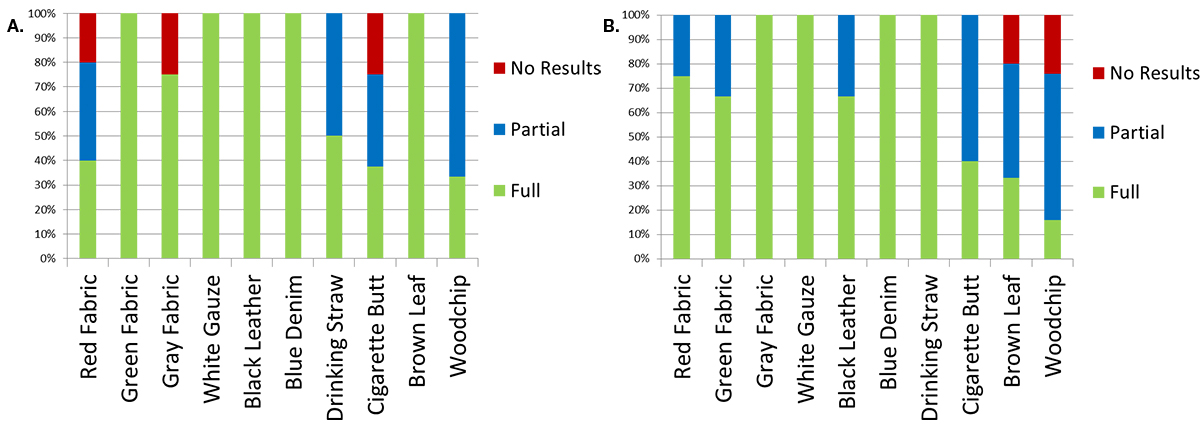 Figure 3. Results of direct amplification.
Figure 3. Results of direct amplification. Panel A. Results of direct amplification of bloodstains from both M1 and M2 donors, deposited on ten substrates using PowerPlex® 18D System. Panel B. Results of direct amplification of bloodstains from both M1 and M2 donors deposited on ten substrates using PowerPlex® Fusion System.
Conclusion
At this time, it is necessary for the forensic community to quantify extracted DNA from evidence samples as dictated by the FBI Quality Assurance (QAS) guidelines. However, with the advent of instruments such as the RapidHIT™ System from IntegenX and the DNAscan™ Rapid DNA Analysis™ System from NetBio and direct amplification of body fluids found on substrates such as the ones used in this research, it might be possible in the near future to directly amplify body fluids identified on crime scene substrates. In conclusion, each laboratory should perform its own internal validation prior to implementing direct amplification from substrates commonly found at crime scenes.
Article References
- Krenke, B.E. et al. (2002) Validation of a 16-locus fluorescent multiplex system. J. Forensic Sci. 47, 773–85.
- Collins, P.J. et al. (2004) Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338, D19S433, and amelogenin: The AmpFlSTR® Identifiler® PCR Amplification Kit. J. Forensic Sci. 49, 1265–77.
- Ensenberger, M.G. et al. (2010) Developmental validation of the PowerPlex 16 HS System: An improved 16-locus fluorescent STR multiplex. Forensic Sci. Int. Genet. 4, 257–64.
- Hall, D.E. and Roy, R. (2014) An evaluation of direct PCR amplification. Croat. Med. J. 55, 655–61.
- Altshuler, H. and Roy, R. (2015) Evaluation of direct PCR amplification using various swabs and washing reagents. J. Forensic Sci. 60, 1542–52..
- Linacre, A. et al. (2010) Generation of DNA profiles from fabrics without DNA extraction. Forensic Sci. Int. Genet. 4, 137–41.
- Larkin, A. and Harbison, S. (1999) An improved method for STR analysis of bloodstained denim. Int. J. Legal Med. 112, 388–90.
- Dargay, A. and Roy, R. (2016) Direct Y-STR amplification of body fluids deposited on commonly found crime scene substrates Journal of Forensic and Legal Medicine 39, 50–60.
- DNA Extraction Using prepGEM® Blood ZyGEM Quick-Start Guide, Accessed April 5, 2015, ZyGEM Corp Ltd.
- Quantifiler® Kits User’s Manual (2005) Applied Biosystems..
- PowerPlex® Fusion System Technical Manual , TMD039, Promega Corporation..
- PowerPlex® 18D System Technical Manual , TMD031, Promega Corporation..
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Gigl, K., Dargay, A. and Roy, R. Direct Amplification of Blood Deposited on Substrates Commonly Encountered at Crime Scenes Using the PowerPlex® 18D and PowerPlex® Fusion Systems. [Internet] 2016. [cited: year, month, date]. Available from: https://www.promega.com/es-es/resources/profiles-in-dna/2016/direct-amplification-of-blood-on-substrates-commonly-encountered-at-crime-scenes/
American Medical Association, Manual of Style, 10th edition, 2007
Gigl, K., Dargay, A. and Roy, R. Direct Amplification of Blood Deposited on Substrates Commonly Encountered at Crime Scenes Using the PowerPlex® 18D and PowerPlex® Fusion Systems. Promega Corporation Web site. https://www.promega.com/es-es/resources/profiles-in-dna/2016/direct-amplification-of-blood-on-substrates-commonly-encountered-at-crime-scenes/ Updated 2016. Accessed Month Day, Year.
PowerPlex is a registered trademark of Promega Corporation. PunchSolution and SwabSolution are trademarks of Promega Corporation.
Applied Biosystems, AmpFlSTR, Identifiler and Quantifiler are registered trademarks of Applied Biosystems. DNAscan is a trademark of GE Healthcare companies. FTA is a registered trademark of Flinders Technologies, Pty, Ltd., and is licensed to Whatman. GeneMarker is a registered trademark of SoftGenetics Corporation. prepGEM is a registered trademark of ZyGEM Corporation, Ltd. Rapid DNA Analysis is a trademark of NetBio. RapidHIT is a trademark of IntegenX.