GoTaq® Hot Start Polymerase for Mouse Genotyping
1Promega Corporation and 2Case Western Reserve University
Publication Date: October 2008
Abstract
GoTaq® Hot Start Polymerase was compared with various Taq DNA polymerases in mouse genotyping assays. The advantages of using the GoTaq® Hot Start Polymerase for this application include more robust amplification, reduced allelic dropout, and increased specificity of the amplification products.
Introduction
Animal models are often used to study the genetic and environmental causes of disease and as well as to screen potential drugs that can be used to treat these diseases. Mouse models can be genetically engineered to contain a particular genotype; therefore, accurate genotyping of these animal models is necessary. PCR is a standard procedure used for genotyping due to the ease and speed of the technique. The DNA template is often purified from crude preparations from mouse tails, toe clippings and ear punches. However, DNA polymerases used for amplification can be affected by inhibitors copurifying with the DNA template. These inhibitors can lead to allelic dropout and errors in genotyping that can result in increased time and cost associated with the studies. In this report, Taq DNA polymerases are compared in mouse genotyping assays to demonstrate the advantages of using Promega GoTaq® Hot Start Polymerase (Cat.# M5001) for this application.
Results
We challenged customers to compare their current Taq DNA polymerase (hot start or no) with GoTaq® Hot Start Polymerase by asking them to submit gel electrophoresis images of the PCR products and describe their mouse genotyping assays results.
Mouse genotyping with transgenic mice
Figure 1 shows the results of a mouse genotyping assay comparing GoTaq® Hot Start Polymerase (Panel A) to a competitor Taq DNA polymerase (Panel B). Ten samples were amplified using each polymerase along with a positive control for the transgenic mice. Using the competitor Taq DNA polymerase, 2 of 10 samples failed to amplify, while the others amplified with varying degrees of efficiency. With GoTaq® Hot Start Polymerase, the customer observed a more robust amplification in all samples tested with no allelic dropout.
 Figure 1. Mouse tail genotyping with transgenic mice.
Figure 1. Mouse tail genotyping with transgenic mice.
Panel A. Amplification with GoTaq® Hot Start Polymerase. Panel B. Amplification with competitor Taq DNA polymerase. Lanes 1–6 and 8–11, sample DNA; lane 7, positive control; lane M, marker.
Mouse genotyping with heterozygous mouse
Figure 2 shows the results of a genotyping assay of a heterozygous mouse comparing GoTaq® Hot Start Polymerase to a competitor Taq DNA polymerase. Decreasing concentrations of MgCl2 were tested in order to optimize the amplification reaction conditions. The expected sizes of the amplified fragments for the heterozygote are 1kb and 350bp. Using the competitor Taq DNA polymerase, the 1kb fragments of the heterozygous mouse are barely visible with each concentration of MgCl2 (Lanes 2–4), which could lead to a false determination of the mouse genotype. On the other hand, GoTaq® Hot Start Polymerase with all concentrations of MgCl2 gave robust amplification with no allelic dropout (Lanes 6–8), resulting in a clear-cut determination of the mouse genotype.
 Figure 2. Mouse tail genotyping with a heterozygous mouse.
Figure 2. Mouse tail genotyping with a heterozygous mouse.
Lane M, 1kb ladder; lanes 1–4, Competitor Taq DNA polymerase; Lanes 5–8, GoTaq® Hot Start Polymerase; lanes 1 and 5, no template; lanes 2 and 6, 6µl MgCl2; lanes 3 and 7, 5µl MgCl2; lanes 4 and 8, 4µl MgCl2.
Mouse genotyping with the Nijmegen breakage syndrome (NBS) gene
Figure 3 shows the results of a mouse genotyping assay with the NBS gene comparing GoTaq® Hot Start Polymerase (Panel A) to two non-hot start DNA polymerases (Panels B and C). The NBS gene is a difficult target to amplify for mouse genotyping. DNA was purified from mouse tails using two different methods—an automated purification method (Maxwell® 16, lanes 1 and 2) and a manual purification method (Wizard® SV Genomic DNA Purification System [Cat.# A2360], lanes 3 and 4). Heterozygous (lane 6) and homozygous controls (lanes 5 and 7) are shown. The polymerase used in Panel B resulted in reduced amplification of the control DNAs and failed to amplify one of the sample DNAs. The polymerase used in Panel C showed very poor results, with little to no amplified product obtained with all of the samples. In contrast, GoTaq® Hot Start Polymerase showed significant improvements in amplification of the NBS gene over the two non-hot start polymerases, with strong amplification and no allelic dropout.
 Figure 3. Mouse tail genotyping using the NBS gene.
Figure 3. Mouse tail genotyping using the NBS gene.
Panel A. Amplification with GoTaq® Hot Start Polymerase; Panels B and C. Amplification with non-hot start DNA polymerases. Lane 1 and 2, DNA purified with automated method; lanes 3 and 4, DNA purified using manual method; lane 5, 440bp homozygous (+/+) control; lane 6, heterozygous (+/–) control; lane 7, 310bp homozygous (–/–) control.
Mouse genotyping with the brain-derived neurotrophic factor (BDNF) gene using the GoTaq® Hot Start Master Mix
Figure 4 shows the results of a mouse genotyping assay with the BDNF gene comparing a non-hot start competitor polymerase (lanes 1–5) with the GoTaq® Hot Start Master Mix (Green option: lanes 7–11; Colorless option: lanes 12–16). Heterozygous and homozygous genotype control DNAs were amplified with each polymerase along with two sample DNAs. In all cases, the heterozygous control failed to amplify, likely due to sample DNA degradation. Using the competitor polymerase, numerous nonspecific fragments amplified, making the genotypes difficult to accurately determine. In stark contrast, both the GoTaq® Green and Colorless Hot Start Master Mixes resulted in a significant increase in amplification specificity with single fragments amplified.
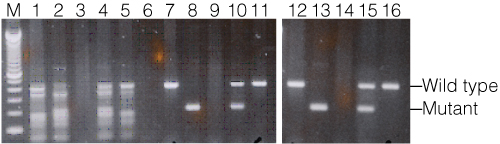 Figure 4. Mouse genotyping with the BDNF gene.
Figure 4. Mouse genotyping with the BDNF gene.
DNA amplified with a standard Taq DNA polymerase (Lanes 1–5), GoTaq® Hot Start Green Master Mix (lanes 7–11), and GoTaq® Hot Start Colorless Master Mix (lanes 12–16). Wild type allele is 400bp, mutant allele is 200bp. Lane M, 100bp DNA ladder; lanes 1, 7, and 12, BDNF (+/+); lanes 2, 8, and 13 BDNF (–/–); lanes 3, 9, and 14, BDNF (+/–); lanes 4, 10, and 15, BDNF mouse colony #646; lanes 5, 11, and 16, BDNF mouse colony #659; lane 6, no-template control
Conclusions
The advantages of using the GoTaq® Hot Start Polymerase and GoTaq® Hot Start Master Mix for mouse genotyping include more robust amplification, reduced allelic dropout, and increased specificity of the amplification products.
Related Products
Related Resources
LabFact #39
For new PCR primers, titrate magnesium in 0.5–1.0mM increments to determine optimum concentration. Some primers require a specific Mg2+ concentration.Related Articles
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Wieczorek, D., Yoder, D., Wang, Q. and Schagat, T. GoTaq® Hot Start Polymerase for Mouse Genotyping. [Internet] October 2008. [cited: year, month, date]. Available from: https://www.promega.com/es-es/resources/pubhub/enotes/gotaq-hot-start-polymerase-for-mouse-genotyping/
American Medical Association, Manual of Style, 10th edition, 2007
Wieczorek, D., Yoder, D., Wang, Q. and Schagat, T. GoTaq® Hot Start Polymerase for Mouse Genotyping. Promega Corporation Web site. https://www.promega.com/es-es/resources/pubhub/enotes/gotaq-hot-start-polymerase-for-mouse-genotyping/ Updated October 2008. Accessed Month Day, Year.
Products may be covered by pending or issued patents or may have certain limitations on use.
 Watch this animated, step-by-step introduction to PCR—a landmark molecular biology technique.
Watch this animated, step-by-step introduction to PCR—a landmark molecular biology technique.