Rapid DNA Digestion Using Promega Restriction Enzymes
Promega Corporation
Publication Date: 2007
Abstract
We tested 30 popular Promega restriction enzymes for their ability to rapidly digest plasmid DNA. To assay enzyme performance, one microliter of each restriction endonuclease was used to cut 1µg of DNA in 5–15 minutes in a total volume of 20µl under standard reaction conditions. Twenty seven of the 30 enzymes tested worked well after a 5-minute incubation. We conclude these enzymes can be used for rapid digestion applications (e.g., quick screening of plasmid clones) without the need to purchase specially formulated enzymes or buffers.
Introduction
To save time and increase productivity, scientists need short, simple protocols for routine laboratory tasks. For restriction enzyme digestions, typical reactions use 1µl of enzyme to cut 1µg of DNA for 1–3 hours. With high-quality, efficient restriction enzymes, this incubation time can be significantly shortened, allowing for rapid screening of clones or preparation of digested DNA for downstream applications. This article describes the successful use of 27 of 30 popular Promega restriction enzymes in a simple rapid-digestion protocol.
Materials and Methods
Restriction enzyme digestions were assembled as follows:
| plasmid DNA1 | 1µg |
| restriction enzyme2 | 1µl |
| 10X buffer | 2µl |
| BSA (10µg/µl) | 0.2µl |
| nuclease-free water to final volume | 20µl |
| 1All enzymes were tested with 1µg of plasmid DNA except for AgeI, which was tested using 1µg of Lambda DNA (Cat.# D1501). 2The number of restriction enzyme units added to the digestion reaction varied with concentration and ranged from 5–12 units. |
|
Reactions were assembled on ice in 0.5ml tubes. For each enzyme, three separate 20µl reactions were incubated at 37°C for 5, 10 or 15 minutes then immediately heat-inactivated at 65°C for 15 minutes. The digestions were analyzed by agarose gel electrophoresis. Note: All the enzymes tested have a recommended incubation temperature of 37°C and can be heat-inactivated at 65°C for 15 minutes. Read the technical literature that accompanies each restriction enzyme for incubation and inactivation conditions.
Results and Conclusions
 Figure 1. Rapid digestion of the pSP72 Vector using the restriction enzymes PvuII, SmaI and DdeI.
Figure 1. Rapid digestion of the pSP72 Vector using the restriction enzymes PvuII, SmaI and DdeI.
For each enzyme, the pSP72 Vector was digested for 5, 10 or 15 minutes and the product compared to uncut pSP72 plasmid DNA. Digestions were analyzed on a 0.7% agarose gel prestained with 0.5µg/ml ethidium bromide. Lane M, 1kb DNA Ladder (Cat.# G5711); lanes 1, 5, 9, uncut pSP72 Vector; lanes 2, 6, 10, 5-minute digestion; lanes 3, 7, 11, 10-minute digestion; lanes 4, 8, 12, 15-minute digestion.
The results from the restriction enzymes, PvuII, SmaI and DdeI, are shown in Figure 1. The uncut DNA, pSP72 Vector (Cat.# P2191), was compared to digestions incubated for 5, 10 or 15 minutes. All three enzymes cut the plasmid DNA efficiently in as little as 5 minutes. In the SmaI digests, trace amounts of undigested pSP72 plasmid DNA were visible at 5, 10 and 15 minutes. Figure 2 shows the results from 5-minute digestions of the pGEM®-5Zf(+) Vector (Cat.# P2241) incubated with NotI, NcoI and AluI and the pSP72 Vector cut with EcoRI, BamHI and NdeI. Each digestion was compared to uncut plasmid DNA. All six enzymes efficiently digested the DNA in 5 minutes. While trace amounts of undigested plasmid DNA were visible in the 5-minute digestions with NotI, NcoI and BamHI, the vector appeared completely cut within 5 minutes for AluI, EcoRI and NdeI.
 Figure 2. Comparison of 5-minute digestions using six restriction enzymes to cut either pGEM®-5Zf(+) or pSP72 Vectors.
Figure 2. Comparison of 5-minute digestions using six restriction enzymes to cut either pGEM®-5Zf(+) or pSP72 Vectors.
NotI, NcoI and AluI were used to digest the pGEM®-5Zf(+) Vector while EcoRI, BamHI and NdeI were used to cut the pSP72 Vector. The digestion products were analyzed on a 0.7% agarose gel prestained with 0.5µg/ml ethidium bromide. Lane M, 1kb DNA Ladder (Cat.# G5711).
Results for all restriction enzymes tested are listed in Table 1. Ninety percent of the enzymes tested worked well in the 5-minute rapid-digestion protocol. The enzymes BglII, PvuI and SacI are not recommended for rapid digestion because significant amounts of uncut DNA were present even after a 15-minute incubation. Enzymes that had trace amounts of uncut DNA during any of the three incubation times were noted with an asterisk (Table 1). As shown in Figures 1 and 2, SmaI, NotI, NcoI and BamHI are examples of enzymes that digest well in 5 minutes but have trace amounts of undigested DNA. The ability to detect the trace undigested DNA will depend on the amount of DNA loaded on the gel and the detection method used.
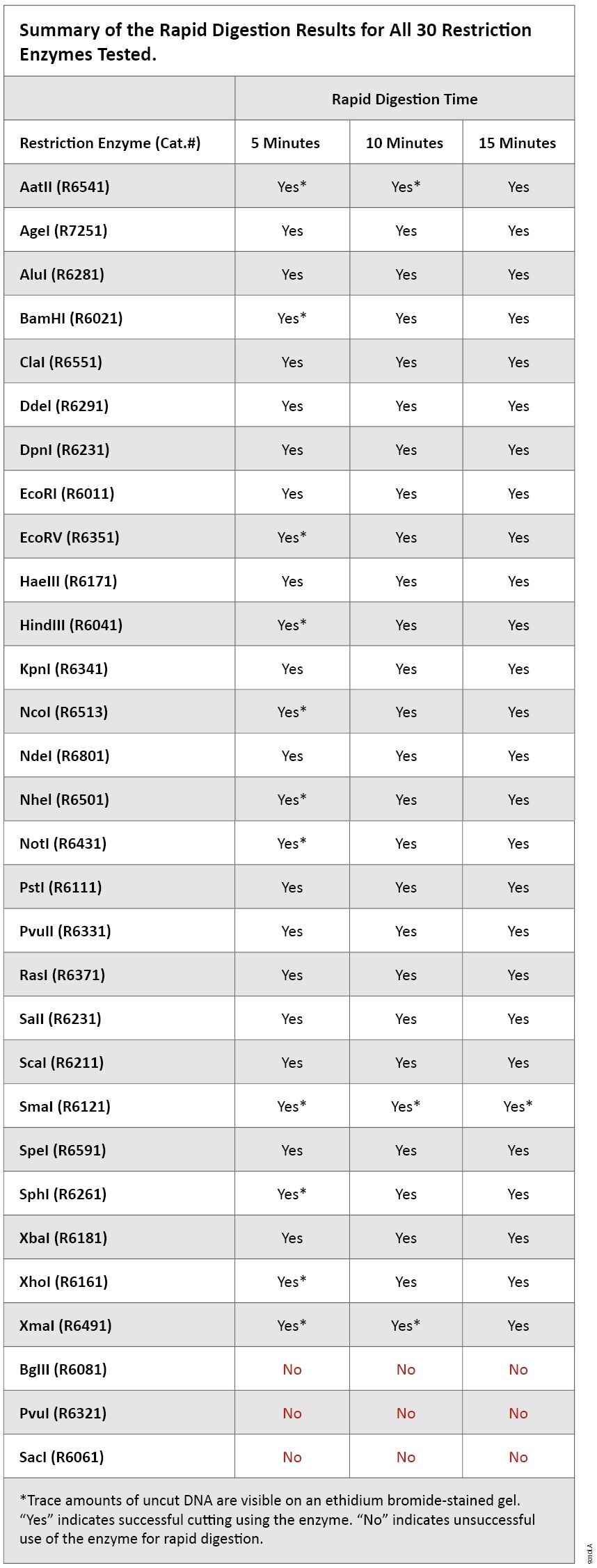 Table 1. Summary of the Rapid Digestion Results for all 30 Restriction Enzymes Tested.
Table 1. Summary of the Rapid Digestion Results for all 30 Restriction Enzymes Tested. The simplicity and convenience of rapid digestion are ideal for screening clones. This method could also be used to prepare digested DNA for other downstream applications, such as ligation. Our TSAP Thermosensitive Alkaline Phosphatase (Cat.# M9910) is active in all restriction endonuclease buffers tested. TSAP dephosphorylates DNA with a 15-minute incubation and could be used with any restriction enzyme to reduce background in subsequent cloning. TSAP is inactivated by heating at 74°C for 15 minutes. Since even trace amounts of undigested DNA can affect cloning by increasing background (i.e., colonies with vector and no insert), gel purification is recommended to remove unwanted DNA products. A rapid purification kit such as the Wizard® SV Gel and PCR Clean-Up System (Cat.# A9281) can be used to extract and purify digested DNA from agarose gel slices in as little as 15 minutes.
Promega restriction enzymes are routinely tested in a number of different quality-control assays including an overdigest assay ( (1) ). The overdigest assay assesses enzyme purity by incubating an excess amount of enzyme with 1µg of DNA for 16 hours. This quality test, in addition to these rapid digestion results, demonstrate these high-quality and efficient restriction enzymes can be used for digesting DNA in as little as 5 minutes or as long as overnight.
Related Products
Related Resources
LabFact #11
New restriction enzyme isoschizomers often are less expensive or offer performance advantages over the classic enzymes. For more information on the use of isoschizomers and neoschizomers, see the Promega Notes article, "Making the Cut...".
Frequently Asked Questions
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Schagat, T. Rapid DNA Digestion Using Promega Restriction Enzymes. [Internet] 2007. [cited: year, month, date]. Available from: https://www.promega.com/es-es/resources/pubhub/enotes/rapid-dna-digestion-using-promega-restriction-enzymes/
American Medical Association, Manual of Style, 10th edition, 2007
Schagat, T. Rapid DNA Digestion Using Promega Restriction Enzymes. Promega Corporation Web site. https://www.promega.com/es-es/resources/pubhub/enotes/rapid-dna-digestion-using-promega-restriction-enzymes/ Updated 2007. Accessed Month Day, Year.
Products may be covered by pending or issued patents or may have certain limitations on use. Please visit our patent and trademark web page for more information.