Titration of Tomato Leaf Material Used as Starting Material for SV Total RNA
Promega Corporation
Publication Date: 1999
Abstract
This article details experiments aimed at demonstrating how much plant material (tomato leaf) can be used for total RNA isolation using the SV Total RNA Isolation System and also to determine the linear range of yield with this system. The isolated total RNA was analyzed by spectrophotometry and reverse transcriptase-PCR (RT-PCR).
Introduction
This article details experiments aimed at demonstrating how much plant material (tomato leaf) can be used for total RNA isolation using the SV Total RNA Isolation System and also to determine the linear range of yield with this system. The isolated total RNA was analyzed by spectrophotometry and reverse transcriptase-PCR (RT-PCR).
Experimental Conditions
RNA Extraction: Frozen tomato leaf tissue (940mg) was ground in liquid nitrogen using a mortar and pestle. Initially, 500mg of tissue was solubilized in 1.3ml SV RNA Lysis Buffer. To this lysate, SV RNA Dilution Buffer (2.6ml) was added. The samples were centrifuged for 10 minutes at 14,000 x g, and ethanol (1.2ml) was added to the supernatant. Volumes of lysate corresponding to 30, 100, 200 and 500mg of original tissue mass were passed through the SV RNA Spin Basket membrane and processed as recommended(1). Note that multiple spins were required to pass the 200mg and 500mg samples through a single Spin Basket membrane; no clogging of the Spin Basket membrane was observed.
RT-PCR Amplification: RNA was next analyzed by RT-PCR using primer pairs specific for the RUBISCO (ribulose 1,5-bisphosphate carboxylase/oxygenase) small subunit gene (rbcS3A) mRNA, a common and abundant plant RNA. Oligonucleotide primers were chosen within highly conserved sequences of the rbcS3A gene.
The primer sequences used were:
- Upstream: 5´-CCCTTCACTGGACTCAAGTCCACCGC-3´
- Downstream: 5´-GCTTGTAAGCGATGAAGCTGATGCACTGC-3´
Each total RNA sample (1µl from a total of 100µl) was reverse transcribed and amplified using the Access RT-PCR System (Cat.# A1250, A1280). The cycling profile was as follows:
| First Strand cDNA Synthesis | |
| 1 cycle | 48°C for 45 minutes |
| 1 cycle | 94°C for 2 minutes |
| Second Strand cDNA Synthesis and PCR Amplification | |
| 35 cycles | 94°C for 30 seconds 60°C for 1 minute 68°C for 1 minute |
| 1 cycle | 68°C for 7 minutes |
| 1 cycle | 4°C soak |
After amplification, the products (10µl) were analyzed on a 1.5% agarose gel (Figure 1).
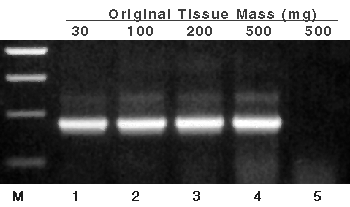 Figure 1. RNA isolated from titrated amounts of starting tomato leaf and used as template in RT-PCR.
Figure 1. RNA isolated from titrated amounts of starting tomato leaf and used as template in RT-PCR.
Aliquots (1µl of 100µl) of the RNA isolated from the indicated tissue masses were used as template in RT-PCR. After amplification, 10µl (one-fifth) of each reaction were separated by agarose gel electrophoresis. Lane 1, 30mg tomato leaf; lane 2, 100mg tomato leaf; lane 3, 200mg tomato leaf; lane 4, 500mg tomato leaf; and lane 5, 500mg tomato leaf without reverse transcriptase in the RT-PCR reaction. Lane M, Promega 100bp DNA Ladder (Cat.# G2101); bands in lane M correspond to 500, 400, 300 and 200bp. Primers for RT-PCR are to the ribulose 1,5-bisphosphate carboxylase/oxygenase (RUBISCO) gene.
Results
The purity of the tomato leaf RNA isolated with the SV Total RNA Isolation System was high, as measured by UV absorbance ratios. Pure preparations of RNA have an A260/A280 ratio of approximately 2.0. If there is protein contamination in the sample, this ratio will be significantly lower(2). A260/A230 ratios lower than 1.5 generally indicate carryover contamination by guanidine thiocyanate. The A260/A230 ratios for the tomato leaf RNA samples isolated with this system were greater than or equal to 2.2, and along with the A260/A280 ratios, indicate high purity of the RNA Table 1).
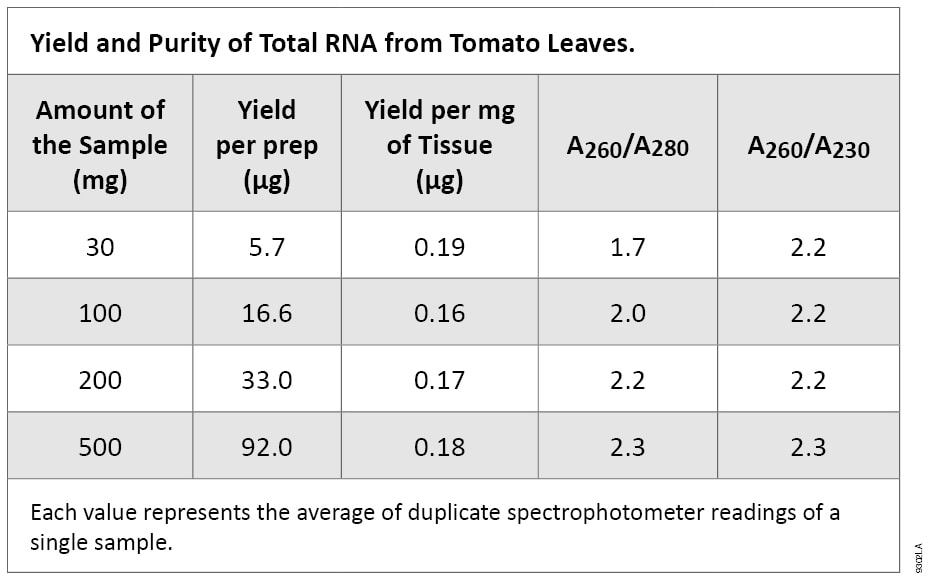 Table 1. Yield and Purity of Total RNA from Tomato Leaves.
Table 1. Yield and Purity of Total RNA from Tomato Leaves. Each value represents the average of duplicate spectrophotometer readings of a single sample.
RNA was isolated from tomato leaf, yielding RNA that was free of detectable genomic DNA in PCR analysis (without reverse transcriptase added) (Figure 1, lane 5). The negative control amplification, which contained RNA from 500mg of tissue without AMV Reverse Transcriptase added, resulted in no product (showing no detectable contaminating genomic DNA.
Summary
The SV Total RNA Isolation System performed well with up to 500mg of tomato leaf material per preparation. The amount of nucleic acid isolated from a sample was linear over the range tested (30mg to 500mg), and spectrophotometric analysis indicated that the RNA is of high quality. All of the samples performed well in RT-PCR amplifications using the RUBISCO primer pair, and there was no detectable genomic DNA in the preparations.
Related Products
Related Protocols
Related Resources
Article References
- SV Total RNA Isolation System Technical Manual, TM048, Promega Corporation.
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Shenoi, H. and Kephart, D. Titration of Tomato Leaf Material Used as Starting Material for SV Total RNA. [Internet] 1999. [cited: year, month, date]. Available from: https://www.promega.com/es-es/resources/pubhub/enotes/titration-of-tomato-leaf-material-used-as-starting-material-for-sv-total-rna/
American Medical Association, Manual of Style, 10th edition, 2007
Shenoi, H. and Kephart, D. Titration of Tomato Leaf Material Used as Starting Material for SV Total RNA. Promega Corporation Web site. https://www.promega.com/es-es/resources/pubhub/enotes/titration-of-tomato-leaf-material-used-as-starting-material-for-sv-total-rna/ Updated 1999. Accessed Month Day, Year.