Recombinant PNGase F for Glycoprotein Analysis
Chris Hosfield1, Laurie Engel1, Aileen Paguio1, Teresa Surowy1, Richard Jones2, Michael Ford2, Marjeta Urh1, Michael Rosenblatt1
1Promega Corporation, Madison, WI; 2MS Bioworks, LLC, Ann Arbor, MI.
Publication Date: 2013
Abstract
PNGase F is an endoglycosidase that specifically removes N-linked glycans from glycoproteins. It is used extensively in workflows for characterizing N-linked glycan structures on therapeutic proteins and for identifying N-linked glycosylation sites in proteomic studies. We have expressed and purified recombinant PNGase F and shown that it is highly active against a variety of substrates, including purified glycoproteins and complex mixtures of human serum glycoproteins. Furthermore, the enzyme is compatible with the mass spectrometry workflow environment, including non-denaturing conditions and the presence of ProteaseMAX™ Surfactant, Trypsin Enhancer. Finally, we show that recombinant PNGase F can be used to functionally deglycosylate therapeutic antibodies as measured by decreased antibody-dependent cell-mediated cytotoxicity after deglycosylation.
Introduction
Glycosylation is among the most abundant of all post-translational modifications (PTM) (1). A highly complex PTM, glycosylation has diverse roles that affect both protein function in cellular processes as well as disease manifestation (2). Several large glycoproteomics studies published recently illustrate how PNGase F can be used in combination with mass spectrometry to identify hundreds to thousands of N-linked glycosylation sites in complex biological samples (2).
Furthermore, glycosylation has been shown to dramatically affect properties of biotherapeutics. For example, the pharmacokinetics and potency of recombinant erythropoietin are critically affected by its glycosylation status, while the ability of monoclonal antibodies to mediate efficacy via ADCC is affected by the fucose content of the Fc region glycan (3). Clearly, with glycosylation having such prominent roles in biology, disease progression and therapeutic protein function, as well as serving as a biomarker in diseases like cancer, tools that facilitate glycoprotein characterization are extremely important.
One such tool is the glycosidase PNGase F, which is used extensively for studying glycoproteins. First isolated from Elizabethkingia miricola (formerly Flavobacterium meningosepticum), PNGase F catalyzes the cleavage of N-linked oligosaccharides between the innermost GlcNAc and asparagine residues of high mannose, hybrid and complex oligosaccharides from N-linked glycoproteins (4). Despite being active against most N-linked glycans, PNGase F cannot cleave when the innermost GlcNAc is linked to an α-1,3-fucose, most commonly found in plant glycoproteins and in some insect glycoproteins (5). During glycan removal, the modified asparagine residue is converted to aspartic acid, which imparts a +1Da mass change that can be detected by mass-spectrometry workflows utilizing trypsin or alternative proteases.
These PNGase F properties make it a valuable scientific tool for use in conjunction with downstream analytical techniques, both to identify specific Asn residues that undergo N-linked glycosylation as well as to characterize the chemical structure(s) of the released oligosaccharide chains.
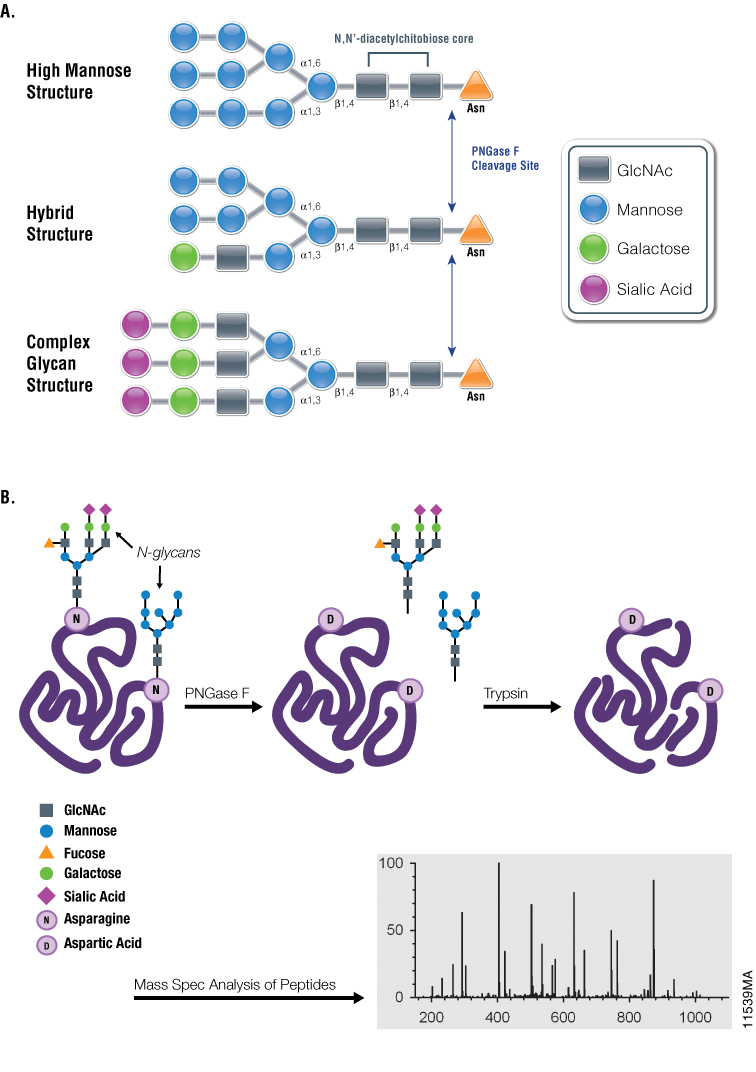
Figure 1. Diagrams showing PNGase F cleavage specificity and its use with mass spectrometry in analysis of N-glycosylation. Panel A. Cleavage specificity of PNGase F. Panel B. Asn-linked glycans can be cleaved by PNGase F, yielding oligosaccharides and a modified protein in which Asn residues, at the site of deglycosylation, are converted to Asp. The conversion of Asn to Asp results in a monoisotopic mass shift of +1Da that can be measured readily using mass spectrometry.
Recombinant PNGase F is Active Against Numerous Substrates
Here we describe a recombinant PNGase F (Cat.# V4831) that has been purified from E. coli and is equally active as the native enzyme. Recombinant PNGase F is slightly larger than the native enzyme due to the presence of an affinity tag used in the purification process.
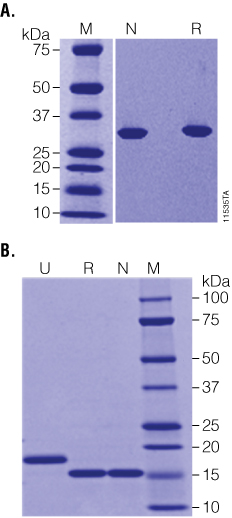
Figure 2. Recombinant PNGase F is highly purified and highly active. Panel A. Native (N) and Recombinant (R) PNGase F are equally pure as determined by SDS-PAGE. Panel B. RNase B was incubated with either native (N) or recombinant (R) PNGase F at high substrate:enzyme ratios and was fully deglycosylated in 1 hour as indicated by the shift from ~18 to 15kDa on SDS-PAGE. Lane M: Precision Plus Protein™ Dual Color Standards (BioRad Cat.# 161-0374).
The enzymatic activity of PNGase F was characterized using the glycoprotein RNase B, a standard substrate often used to measure PNGase F activity. Even at very high ratios of substrate:enzyme (20µg RNase B:1 unit (0.1µl) of recombinant PNGase F; as shown in Figure 2, Panel B), recombinant PNGase F deglycosylates RNase B as effectively as the native enzyme in 1 hour at 37°C, indicating that the affinity tag does not adversely affect the enzyme activity. Recombinant PNGase F was also tested against a wider panel of glycoproteins. As seen in Figure 3, Panel A, 20µg of various glycoproteins were completely deglycosylated after a 2-hour treatment with 20 units of recombinant PNGase F at 37°C.
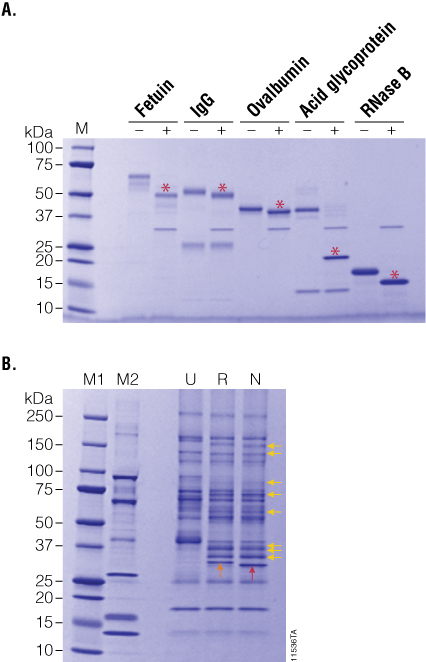
Figure 3. PNGase F treatment of purified and serum-enriched glycoproteins. Panel A. Purified glycoproteins were incubated in the presence (+) or absence (–) of recombinant PNGase F for 2 hours. Red asterisks indicate the deglycosylated product. Panel B. Enriched glycoproteins from human serum were incubated with either recombinant (R) or native (N) PNGase F, and the deglycosylation patterns (yellow arrows) were analyzed by SDS-PAGE. Lane M1: Precision Plus Protein™ Dual Color Standards (BioRad Cat.# 161-0374). Lane M2: CandyCane™ Glycoprotein Molecular Weight Standards (Life Technologies Cat.# C-21852).
To further compare the activity profiles of native and recombinant PNGase F, we prepared a complex sample of lectin-enriched glycoproteins from human serum. Briefly, human serum was depleted of albumin and IgG, and the resultant sample was enriched for serum glycoproteins using immobilized lectins ConA and WGA. After elution from the lectin columns, glycoproteins were desalted by ultrafiltration, and 10µg was treated with 5 units of recombinant PNGase F overnight at 37°C. The resultant glycosylation profiles on SDS-PAGE indicate that both native and recombinant PNGase F perform equally well on complex samples (Figure 3, Panel B).
Using Recombinant PNGase F in Mass Spectrometry-Compatible Conditions
PNGase F is often most effective at deglycosylating protein substrates that are first denatured; typically with detergents and reducing agents. Denatured substrates provide PNGase F with efficient access to the oligosaccharide-protein linkage. However, since PNGase F is often used in workflows that involve downstream characterization of the liberated protein or glycans using downstream proteolysis and mass spectrometry, it would be advantageous to be able to perform deglycosylation reactions under non-denaturing, mass spectrometry-compatible conditions. To test whether recombinant PNGase F was active under such conditions, we incubated the enzyme with a variety of glycoprotein substrates in ammonium bicarbonate buffer lacking denaturants. As indicated in Figure 4, while many of the glycoprotein substrates tested showed reduced deglycosylation under non-denaturing conditions, the heavy chain of human IgG could be fully deglycosylated. Although not fully deglycosylated under the tested conditions, fetuin was partially deglycosylated by PNGase F. Interestingly, RNase B, which is an excellent substrate under denaturing conditions, is quite resistant to PNGase activity in native conditions, while ovalbumin was fully resistant.
We tested the ability of ProteaseMAX™ Surfactant, Trypsin Enhancer, (Cat.# V2071) to improve deglycosylation under mass spectrometry-compatible conditions. In these experiments, 20µg of various glycoproteins was incubated with 20 units of recombinant PNGase F and ProteaseMAX™ Surfactant ranging from 0 to 0.1% for 90 minutes at 37°C. As shown in Figure 4, addition of ProteaseMAX™ Surfactant significantly improved the ability of recombinant PNGase F to deglycosylate the three proteins that were not completely deglycosylated under native conditions. In the presence of ProteaseMAX™ Surfactant, fetuin was fully deglycosylated, while both RNase B and ovalbumin were much more efficiently deglycosylated than compared to controls lacking ProteaseMAX™ Surfactant.
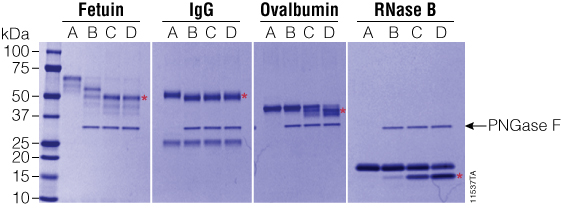
Figure 4. Effect of ProteaseMAX™ Surfactant on PNGase F activity. PNGase F was incubated with various glycoproteins with increasing concentrations of ProteaseMAX™ Surfactant (Cat.# V2071), and deglycosylation was monitored by SDS-PAGE. Red asterisks indicate the deglycosylated products. Gel column contents are as follows: A, no enzyme control; B, native conditions (50mM ammonium bicarbonate buffer); C, 50mM ammonium bicarbonate buffer + 0.05% ProteaseMAX™ Surfactant; D, 50mM ammonium bicarbonate buffer + 0.1% ProteaseMAX™ Surfactant. Lane M: Precision Plus Protein™ Dual Color Standards (Bio-Rad Cat.# 161-0374).
PNGase F and ADCC
N-glycosylation of the Fc region of antibodies is known to be essential for ADCC. To test the ability of PNGase F to functionally deglycosylate therapeutic monoclonal antibodies, we used the ADCC Reporter Bioassay, Complete Kit (WIL2-S) (Cat.# G7014). Rituximab was incubated with recombinant PNGase F under non-denaturing conditions and was fully deglycosylated as indicated by SDS-PAGE. Blends of N-glycosylated and deglycosylated samples were then prepared in various ratios and assayed in the reporter bioassay. As seen in Figure 5, the fully deglycosylated Rituximab had no detectable ADCC, while antibody with even 10% deglycosylation showed measurable activity.
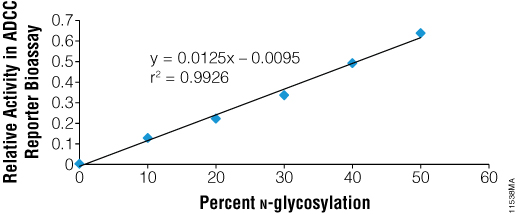
Figure 5. Recombinant PNGase F deglycosylates therapeutic antibodies as shown by ADCC. Rituximab was deglycosylated using PNGase F and blended with the fully N-glycosylated parent preparation to create preparations representing different % N-glycosylation. Blended mixtures were assayed together in the ADCC Reporter Bioassay Complete Kit (WIL2-S (Cat.# G7014) and biological activity was determined relative to parent.
Summary
Recombinant PNGase F is a highly active enzyme that can be used to efficiently deglycosylate N-linked glycoproteins for characterization of glycan structure and function, as well as for glycan-site identification. We have demonstrated that recombinant PNGase F expressed in E. coli is equally pure and active compared to the native enzyme, and can be used against a variety of substrates and in multiple applications.
It is ideal for use in mass spectrometry-based workflows, in combination with other enzymes such as trypsin and with mass spectrometry-compatible surfactants, such as ProteaseMAX™ Surfactant.
References
- Khoury, G.A., Baliban R.C., Floudas C.A. (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 1-5.
- Lazar, I.M., Lee, W., Lazar, A.C. (2013) Glycoproteomics on the rise: Established methods, advanced techniques, sophisticated biological applications. Electrophoresis 34, 113-25.
- Li, H., d'Anjou, M. (2009) Pharmacological significance of glycosylation in therapeutic proteins. Curr. Opin. Biotechnol. 20, 678-84.
- Elder, J.H., Alexander, S. (1982) Endo-beta-N-acetylglucosaminidase F: Endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proc. Natl. Acad. Sci. USA 79, 4540-4.
- Tretter, V., Altmann, F., März, L. (1991) Peptide-N4-(N-acetyl-beta-glucosaminyl) asparagine amidase F cannot release glycans with fucose attached α-1,3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199(3), 647-52.