Allele Frequencies for Penta C in Caucasian, African American, American Indian, Asian and Hispanic Populations
Laboratory Corporation of America Holdings, Burlington, North Carolina, USA
Publication Date: 2011
Introduction
The Promega PowerPlex® CS7 System contains seven loci: FES/FPS, LPL, F13A, F13B, Penta D, Penta E and Penta C. All of these loci, with the exception of Penta C, are used in other multiplex systems. As a result, Penta C allele frequencies are not currently available for a diverse set of races. This study presents Penta C allele frequencies for Caucasian, African American, American Indian, Asian and Hispanic populations.
Materials and Methods
PowerPlex® CS7 Systems were obtained from Promega. Extracted DNA was analyzed using the Applied Biosystems 3130xl platforms and GeneMapper® ID software, version 3.2.1. For each population 300 persons from a convenience sample were analyzed. The race of the individuals was self-identified. Results were analyzed for heterozygosity(1), Hardy Weinberg Equilibrium using the Fisher Exact Test with 10,000 shufflings(1), PIC values(2), probability of a match(3), power of discrimination(3), average probability of exclusion(4) and an estimate of the typical paternity index(5).
View a pdf version of Table 1.
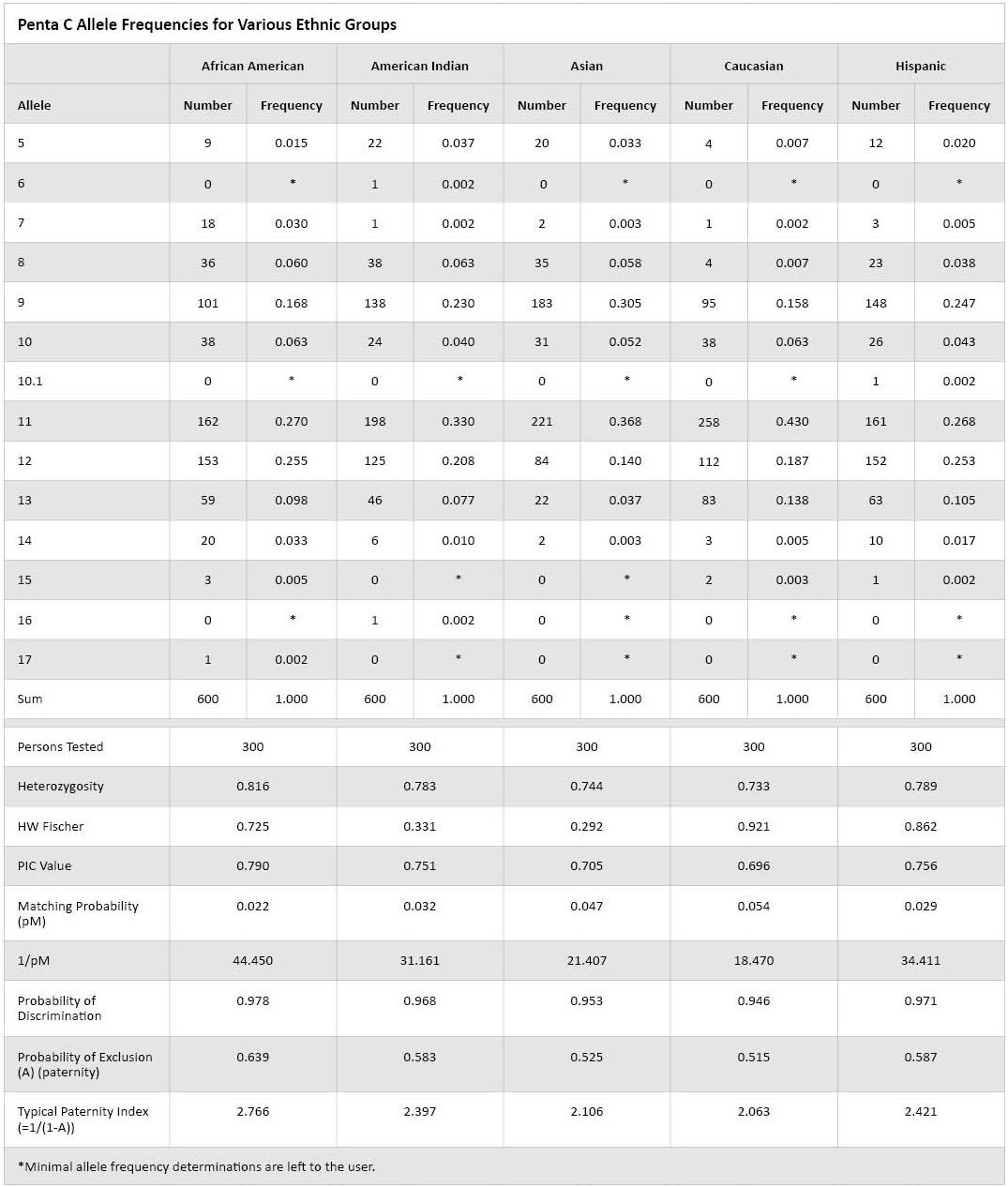 Table 1. Penta C Allele Frequencies for Various Ethnic Groups.
Table 1. Penta C Allele Frequencies for Various Ethnic Groups. The Asian population used in this study consisted primarily of Chinese individuals and other Asian ethnic groups. The Asian data presented are similar to Japanese Penta C frequencies(6). Note that in this table the choice of minimal allele frequencies is left to the laboratory as there are differing methods on the determination of minimal allele frequencies(7)(8). All populations were in Hardy Weinberg Equilibrium using the Fisher Exact Test. The typical paternity index ranged from 2.063 to 2.766, and the match probability ranged from 0.022 to 0.054. In conclusion, Penta C is a valid option for the expansion of current multiplex systems.
Related Products
Article References
- Lewis, P.O. and Zaykin, D. (2001) Genetic data analysis: Computer program for the analysis of allelic data. Version 1.0 (d16c). Free program distributed by the authors at: http://lewis.eeb.uconn.edu/lewishome/software.html.
- Botstein, D. et al. (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–31.
- Huston, K. (1998) Statistical analysis of STR data. Profiles in DNA 1(3), 14–5.
- Garber, R.A. and Morris, J.W. (1983) General equations for the average power of exclusion for genetic systems of n codominant alleles in one-parent and no-parent cases of disputed parentage. In: Inclusion Probability in Parentage Testing, R.H. Walker, ed., American Association of Blood Banks, Arlington, 277–80.
- Morris, J.W. (1983) Relationship between power of exclusion and probability of paternity. In: Inclusion Probability in Parentage Testing, R.H. Walker, ed., American Association of Blood Banks, Arlington, 267–76.
- Ohtaki, H. et al. (2006) Allele frequency distributions at three pentanucleotide repeat STRs in a Japanese population. J. Forensic Sci. 51, 193–4.
- (1996) The evaluation of forensic DNA evidence Committee on DNA Forensic Science: An update, National Research Council, National Academy Press,Washington, D.C.
- Gjertson, D.W. et al. (2007) ISFG: Recommendations on biostatistics in paternity testing. Forensic Sci. Int. Genet. 1, 223–31.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Maha, G.C. and Fuller, J.R. Allele Frequencies for Penta C in Caucasian, African American, American Indian, Asian and Hispanic Populations. [Internet] 2011. [cited: year, month, date]. Available from: https://www.promega.com/resources/profiles-in-dna/2011/allele-frequencies-for-penta-c/
American Medical Association, Manual of Style, 10th edition, 2007
Maha, G.C. and Fuller, J.R. Allele Frequencies for Penta C in Caucasian, African American, American Indian, Asian and Hispanic Populations. Promega Corporation Web site. https://www.promega.com/resources/profiles-in-dna/2011/allele-frequencies-for-penta-c/ Updated 2011. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
PowerPlex is a registered trademark of Promega Corporation.
GeneMapper is a registered trademark of Life Technologies.