PowerPlex® ESX and ESI Systems Technical Tips
Promega Corporation
Publication Date: 2010
The five-dye PowerPlex® ESX and ESI Systems each contain the five new loci selected by the European Network of Forensic Science Institutes (ENFSI) and European DNA Profiling (EDNAP) Group (D10S1248, D22S1045, D2S441, D12S391 and D1S1656) as well as 11 other loci commonly used throughout Europe (D8S1179, D18S51, D21S11, FGA, TH01, vWA, D2S1338, D3S1358, D16S539, D19S433 and the sex-determining marker Amelogenin). The PowerPlex® ESX Systems deliver the D10S1248, D22S1045 and D2S441 loci as miniSTRs (<125bp) and D12S391 and D1S1656 as midiSTRs (125–185bp). The PowerPlex® ESI Systems include the new ENFSI/EDNAP loci but focus on miniaturization of current European Standard Set (ESS) loci: Amplification products for seven of the ten commonly used European loci, including D3S1358, D19S433, D16S539, D18S51, D8S1179, TH01 and vWA, are all less than 230 bases in length. The smaller FGA alleles also are less than 230 bases. Both the ESX and ESI formats are available with and without SE33.
Which dye set and run module do I use for PowerPlex® 5-dye matrix generation on my ABI PRISM® instrument?
Dye sets and run modules vary with the instrument type and data collection software version used. See Table 1.
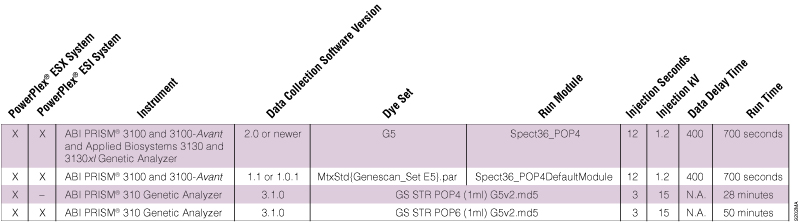 Table 1. Dye Sets and Run Modules for the PowerPlex® 5-Dye Matrix Standards on Different ABI PRISM® Instruments.
Table 1. Dye Sets and Run Modules for the PowerPlex® 5-Dye Matrix Standards on Different ABI PRISM® Instruments. Can I use the same run modules and injection settings for both matrix generation and sample analysis?
No, the run modules and injection settings used for sample runs differ from those used for matrix generation. See Table 2.
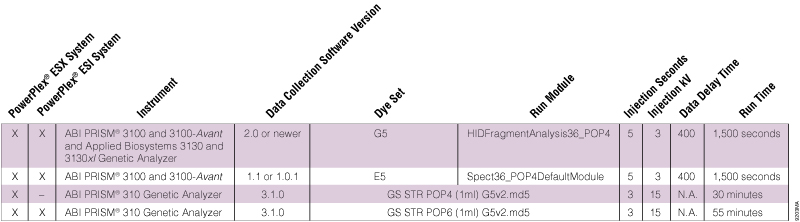 Table 2. Dye Sets and Run Modules for PowerPlex® ESX and ESI Sample Analysis on Different ABI PRISM® Instruments.
Table 2. Dye Sets and Run Modules for PowerPlex® ESX and ESI Sample Analysis on Different ABI PRISM® Instruments. The PowerPlex® ESI Allelic Ladder fragments analyzed using my ABI PRISM® 310 Genetic Analyzer are not assigned properly. Allele calls at D1S1656, D2S441 and/or D12S391 are missing (Figure 1). What can I do?
Poor resolution of alleles, especially those that are 1 base apart in larger loci, results in fewer peaks than the GeneMapper® ID software expects at the locus. As a consequence, the software does not label peaks at this locus and does not shift the location of bins to match the observed sizing of alleles at that locus. Figure 2 shows examples of good and poor resolution of alleles in the D1S1656 allelic ladder.
 Figure 1. Missing allele calls at D1S1656, D2S441 and D12S391 loci.
Figure 1. Missing allele calls at D1S1656, D2S441 and D12S391 loci.  Figure 2. Resolution of D1S1656 allelic ladder fragments.
Figure 2. Resolution of D1S1656 allelic ladder fragments.
Panel A. Poor resolution of D1S1656 allelic ladder fragments. Alleles are not assigned. Panel B. Good resolution of D1S1656 allelic ladder fragments. Alleles are called correctly.
I do not have the GS STR POP6 (1ml) G5v2.md5 module installed on my ABI PRISM® 310 Genetic Analyzer. What can I do?
You can easily create the GS STR POP6 (1ml) G5v2.md5 module by following the instructions below.
- Open the ABI PRISM® 310 Data Collection Software, Version 3.1.0.
- Select "Manual Control" in the Window menu.
- Choose the GS STR POP4 (1ml) G5v2.md5 module using the pull-down menu under "Module".
- Click on the folded page icon (Figure 3).
- Change the Collection Time to "50" and Syringe Pump Time to "360" (Figure 4). Select "Save Copy in".
- Save the new module in the Modules folder. Change the file name to GS STR POP6 (1ml) G5v2.md5, and select "Save" (Figure 5).
 Figure 3. The Manual Control screen.
Figure 3. The Manual Control screen.  Figure 4. Changing the collection time and syringe pump time.
Figure 4. Changing the collection time and syringe pump time.  Figure 5. The Save screen.
Figure 5. The Save screen. When using the Applied Biosystems 3130 or 3130xl Genetic Analyzer, I see imbalance across the dyes in my PowerPlex® ESX or ESI DNA profiles, with the blue and green signals more dominant than yellow and red signals. What can I do?
If the balance of the allelic ladder profile also is poor, double-check that you used the correct dye set and run module to generate the matrix. Be sure to use the recommended G5 dye set; use of "Any 5 Dye" as the dye set during capillary electrophoresis will result in yellow and red signals being lower than blue and green signals.
If the balance of the allelic ladder profiles looks good and only the sample profiles are imbalanced, consult the troubleshooting steps below.
1. Double-check your DNA elution buffer.
We recommend the use of TE–4 Buffer to elute DNA samples. If you are using the "traditional" TE Buffer, which has a higher EDTA concentration, the EDTA can chelate Mg2+ ions in the amplification reaction. The DNA polymerase will be negatively affected by the lower Mg2+ concentration, resulting in poor amplification of fragments labeled with the red dye.
Compare the compositions of TE–4 Buffer and TE Buffer:
| TE–4 Buffer | TE Buffer |
| 10mM Tris-HCl | 10mM Tris-HCl |
| 0.1mM EDTA (pH 8.0) | 1mM EDTA (pH 8.0) |
2. Double-check the thermal cycling protocol.
We recommend the following protocol for the GeneAmp® PCR System 9700 and 2720 thermal cyclers:
96°C for 2 minutes, then
94°C for 30 seconds
59°C for 2 minutes
72°C for 90 seconds
for 30 cycles, then
60°C for 45 minutes
4°C soak
Note: For optimal results, the thermal cycler must accurately heat and cool reactions to the programmed temperatures. Annealing temperatures higher than 59°C can cause imbalance of your profiles. In particular, the D8S1179 and FGA loci are sensitive to higher annealing temperatures and will exhibit a dramatic decrease in peak heights with a 2°C increase in annealing temperature. In the PowerPlex® ESI system, both of these loci are in the red channel. To minimize temperature inaccuracies during PCR cycling, calibrate your thermal cyclers on a routine basis.
How can I improve resolution of allele fragments?
There are several things that you can do to improve resolution.
- Do not overload the capillary. Samples with higher peak heights will show lower resolution. Allelic ladder peak heights should be about 1,000RFU.
- Perform maintenance procedures on a routine basis and replace the capillary in a timely manner to ensure good running conditions on your ABI PRISM® 310 Genetic Analyzer. We recommend changing polymer and buffer every week.
- Use performance optimized polymer 6 (POP-6™). Resolution of alleles with POP-6™ polymer is much better than that with POP-4™ polymer. Using POP-6™ will result in better separation of larger fragments that differ by only 1 bp in length and better allele calling by GeneMapper® ID or Genotyper® software. POP-6™ polymer might be necessary when using the PowerPlex® ESI System to resolve alleles 17.3 and 18, and 18.3 and 19 in the D12S391 allelic ladder, alleles 11.3 and 12 in the D2S441 allelic ladder and alleles 14.3 and 15, 15.3 and 16, 16.3 and 17, 17.3 and 18, 18.3 and 19 in the D1S1656 allelic ladder.
- As an interim solution, consider reducing the Peak Window Size to allow the software to detect peaks that are poorly resolved. This can be done by modifying the Peak Window Size in the Analysis Method Editor to help improve resolution (Figure 6). The default value for Peak Window Size is 15 pts, but this value can be reduced incrementally to 5 pts if necessary.
 Figure 6. The Analysis Method Editor window.
Figure 6. The Analysis Method Editor window. Related Articles
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Weispfenning, R. PowerPlex® ESX and ESI Systems Technical Tips. [Internet] 2010. [cited: year, month, date]. Available from: https://www.promega.com/resources/profiles-in-dna/powerplex-esx-and-esi-systems-technical-tips/
American Medical Association, Manual of Style, 10th edition, 2007
Weispfenning, R. PowerPlex® ESX and ESI Systems Technical Tips. Promega Corporation Web site. https://www.promega.com/resources/profiles-in-dna/powerplex-esx-and-esi-systems-technical-tips/ Updated 2010. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
PowerPlex is a registered trademark of Promega Corporation.
ABI PRISM, GeneMapper and Genotyper are registered trademarks of Applera Corporation. GeneAmp is a registered trademark of Roche Molecular Systems, Inc. POP-4 and POP-6 are trademarks of Applera Corporation.
Products may be covered by pending or issued patents or may have certain limitations. More information.
All prices and specifications are subject to change without prior notice.
Product claims are subject to change. Please contact Promega Technical Services or access the Promega online catalog for the most up-to-date information on Promega products.