Activity of Promega Restriction Enzymes in GoTaq® Green and PCR Master Mixes
Promega Corporation
Publication Date: 2006
Abstract
Minimizing the steps needed to analyze a PCR product saves researchers both time and reagents. In this experiment, 37 restriction enzymes were tested for activity in Promega GoTaq® Green Master Mix or PCR Master Mix. The results demonstrated 32 enzymes cut equivalently to parallel digests in recommended restriction buffers, while the five remaining exhibited star activity.
Introduction
Many applications exist that require restriction digests of PCR products prior to cloning or analysis. Two such applications are restriction fragment length polymorphism (RFLP) genotyping (1)(2) and single-stranded conformational polymorphism (SSCP) analysis. While PCR products can be purified prior to restriction endonuclease digestion, you can save both time and reagents by performing restriction digests in the same buffer conditions as the amplification reaction. We present data reporting the activity of Promega restriction enzymes in both our GoTaq® Green Master Mix and PCR Master Mix.
As the following results indicate, these restriction enzymes can be used after amplification without having to perform an extra purification step. Eliminating a step reduces potential pipetting error and decreases assay preparation time especially for high-throughput applications. Furthermore, use of GoTaq® Green Master Mix eliminates the need to add gel loading buffer, which removes an additional step prior to electrophoresis.
Materials and Methods
In these experiments, the restriction enzyme was added directly to 1X Master Mix. For each restriction enzyme, the Lambda DNA (Cat.# D1501) was digested in each of three reactions:
Standard Buffer Conditions (positive control)
| nuclease-free water | 21.25µl |
| 100X BSA | 0.25µl |
| 500ng Lambda DNA | 1µl |
| Master Mix Volume | 22.5µl |
For Each Reaction
| master mix | 22.5µl/rxn |
| 10X digestion buffer | 2.5µl |
| restriction enzyme | 0.5µl |
| Total Volume | 25.5µl |
1X GoTaq® Green Master Mix
| 2X GoTaq® Green Master Mix | 12.5µl |
| 500ng Lambda DNA | 1µl |
| nuclease-free water | 11µl |
| Master Mix Volume | 25µl |
For Each Reaction
| master mix | 25µl/reaction |
| restriction enzyme | 0.5µl |
| Total Volume | 25.5µl |
1X PCR Master Mix
| 2X PCR Master Mix | 12.5µl |
| 500ng Lambda DNA | 1µl |
| nuclease-free water | 11µl |
| Master Mix Volume | 25µl |
For Each Reaction
| master mix | 25µl/rxn |
| restriction enzyme | 0.5µl |
| Total Volume | 25.5µl |
All digestion reactions were incubated for 2 hours at 37°C in a plate incubator. After digestion, the samples were loaded onto 0.8% agarose gels to monitor cutting efficiency. Two no-restriction-enzyme controls with either 1X GoTaq® Green Master Mix or 1X PCR Master Mix were loaded in the leftmost lanes of each gel. Five microliters of the 6X Blue/Orange Loading Dye (Cat.# G1881) was added to each of the standard restriction enzyme digests before electrophoresis.
The agarose gels were run at 50 volts for ~1.5 hours and stained using 0.5µg/ml ethidium bromide solution with shaking for 15 minutes followed by destaining with shaking for 15 minutes. The stained bands were visualized on a UV light box and photographed with Polaroid® 667 black and white film.
Results
As expected, there was no digestion of template DNA in the absence of restriction enzyme in either GoTaq® Green Master Mix or in PCR Master Mix (wells 1 and 2 in Figure 1, Panels A and B). Of the 37 enzymes tested for activity in GoTaq® Green Master Mix and PCR Master Mix, 32 showed digestion comparable to the standard restriction enzyme digestion conditions with the supplied buffer. These results indicate that 0.5µl volume of restriction enzyme can be added directly to the completed PCR without need for dilution or transfer to a separate tube (Figure 1, Panel A, and Table 1). The remaining 5 restriction enzymes (AatII, BalI, EcoRI, ScaI and XhoII) showed unacceptable deviation from the standard digestion pattern, likely due to star activity. Therefore, these five enzymes are not recommended for direct digestion of PCR products (Figure 1, Panel B).
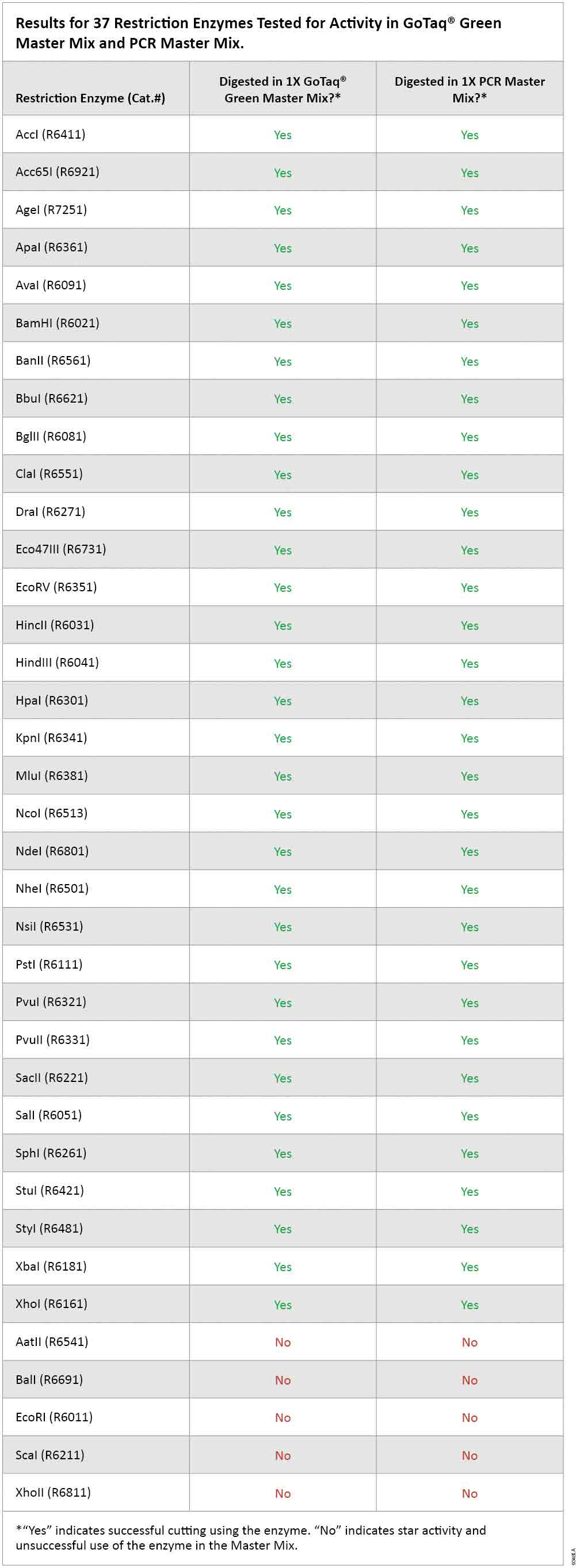 Table 1. Results for 37 Restriction Enzymes Tested for Activity in GoTaq® Green Master Mix and PCR Master Mix.
Table 1. Results for 37 Restriction Enzymes Tested for Activity in GoTaq® Green Master Mix and PCR Master Mix. Conclusions
The results reported here using Lambda DNA template indicate that restriction enzyme digestion could be performed successfully on PCR or RT-PCR products in GoTaq® Green Master Mix or PCR Master Mix without purification. Our data indicate complete digestion can be attained simply by adding restriction enzyme directly into the PCR cocktail and incubating at 37°C with no need for further manipulation. Of the 37 enzymes tested, 32 showed digestion qualitatively similar to that achieved in the supplied standard buffer, while the remainder showed star activity.
This streamlined method could facilitate RFLP analyses if performed directly in Promega PCR master mixes. Another potential application would be a one-step cloning without the need for separate restriction digestion and gel purification of the amplified insert. One caveat is the suboptimal activity of thermostable DNA polymerases at 37°C (3). Even after thermal cycling, the thermostable DNA polymerase retains activity, which could fill in the sticky ends generated during RE digestion. These blunt-ended products can result in reduced cloning efficiency. While adding a restriction enzyme directly to PCR saves time, purifying the DNA product following digestion might be advantageous in removing small restriction fragments that could interfere with ligation.
Related Products
Related Protocols
Related Resources
LabFact #39
For new PCR primers, titrate magnesium in 0.5–1.0mM increments to determine optimum concentration. Some primers require a specific Mg2+ concentration.Article References
- Rodriguez-Nava, R. et al. (2006) Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J. Clin. Microbiol. 44, 536–46.
- Conville, P.S. and Witebsky, F.G. (2005) Multiple copies of the 16S rRNA gene in Nocardia nova isolates and implications for sequence-based identification procedures. J. Clin. Microbiol. 43, 2881–5.
- Turbett, G.V. and Sellner, L.N. (1996) Digestion of PCR and RT-PCR products With restriction endonucleases without prior purification or precipitation. Promega Notes 60, 23–7.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Tritle, D. Activity of Promega Restriction Enzymes in GoTaq® Green and PCR Master Mixes. [Internet] 2006. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/enotes/activity-of-promega-restriction-enzymes-in-gotaq-green-and-pcr-master-mixes/
American Medical Association, Manual of Style, 10th edition, 2007
Tritle, D. Activity of Promega Restriction Enzymes in GoTaq® Green and PCR Master Mixes. Promega Corporation Web site. https://www.promega.com/resources/pubhub/enotes/activity-of-promega-restriction-enzymes-in-gotaq-green-and-pcr-master-mixes/ Updated 2006. Accessed Month Day, Year.
GoTaq is a registered trademark of Promega Corporation.
Polaroid is a registered trademark of Polaroid Corporation.
Products may be covered by pending or issued patents or may have certain limitations. Please visit our Web site for more information.
 Watch this animated, step-by-step introduction to PCR—a landmark molecular biology technique.
Watch this animated, step-by-step introduction to PCR—a landmark molecular biology technique.