Bioluminescent Reporter Genes
Promega Corporation
Publication Date: October 2008
Abstract
Reporter genes typically are used to monitor changes in transcriptional rate, which can vary quickly in response to a specific cellular event. Here we give background on bioluminescent reporters and assays and their uses in research.
Introduction
Bioluminescence is a special form of chemiluminescence found in living organisms. This class of chemiluminescence is an enzyme-catalyzed process, where the high efficiency of photon emission is derived through natural evolution. The enzymes are called luciferases, and the photon-emitting substrates are luciferins. Bioluminescent chemistries have evolved from multiple independent origins and comprise many distinct molecular structures. Of the many natural forms, two have been used widely for genetic reporter assays: firefly luciferase and Renilla luciferase (1) .
Genetic reporters are used widely as indicators to study gene expression and cellular events coupled to gene expression for pharmaceutical and biomedical research and in molecular biology and biochemistry. Typically, a reporter gene is cloned with a DNA sequence of interest into an expression vector that is then transferred into cells. Following transfer, the cells are assayed for the presence of the reporter by directly measuring the reporter protein itself or the enzymatic activity of the reporter protein. A good reporter gene can be identified easily and measured quantitatively when it is expressed in the organism or cells of interest.
Although a number of reporter technologies have been developed for use in the laboratory, the luciferase reporter assays give the researcher unparalleled sensitivity, dynamic range, versatility and ease of use. In this article, we focus on two luciferase reporters, firefly and Renilla, the development of recombinant proteins and their use in dual-reporter assays, since much of the luciferase reporter gene technology was developed by Promega. We discuss factors in choosing a reporter vector, the advantages of bioluminescence and other uses of luciferase reporters.
The Bioluminescence Advantage
Bioluminescent reporter gene assays have a distinct advantage over fluorescent assays, such as green fluorescent protein (GFP), in that they can deliver 10- to 1,000-fold higher assay sensitivity. This greatly increased sensitivity can substantially improve assay performance when applied to complex biological samples (2) . The reason for this becomes clear when examining the mechanisms of light production by fluorescence and luminescence. Both processes yield photons as a consequence of energy transitions from excited-state molecular orbitals to lower-energy orbitals. However, they differ in how the excited-state orbitals are created. In luminescence, the excited states are the product of exothermic chemical reactions, whereas in fluorescence the excited states are created by absorption of light. In reporter assays, bioluminescence has the advantage because photons are not required to create the excited states. Therefore, they do not constitute an inherent background when measuring photon efflux from a sample. The resulting low background permits precise measurement of small changes in light across four to eight orders of magnitude in the linear range of the assay.
Luciferase Reporters
The luciferase reporter technology is based on the interaction of the enzyme luciferase with a luminescent substrate luciferin, which releases light by the process of bioluminescence. Bioluminescence is found in a number of different organisms; however, it is firefly (Photinus pyralis) luciferase that is by far the most commonly used bioluminescent reporter due to both the sensitivity and convenience of the enzyme assay and to the tight coupling of protein synthesis with enzyme activity. This monomeric enzyme of 61kDa catalyzes a two-step oxidation reaction to yield light, usually in the green to yellow region, typically 550–570nm (Figure 1). Upon mixing with substrates, firefly luciferase produces an initial burst of light that decays over about 15 seconds to a low level of sustained luminescence. The gene encoding firefly luciferase (luc) is a cDNA and does not require any post-translational modifications. This means it is available as a mature enzyme directly upon translation from its mRNA, an important consideration when using a reporter gene outside its native environment.
The other commonly used luciferase is Renilla luciferase from Renilla reniformis, a coelenterate that creates bright green flashes upon tactile stimulation. Renilla luciferase is a 36kDa monomeric enzyme that catalyzes the oxidation of coelenterazine to yield coelenteramide and blue light of 480nm (Figure 1). Like firefly luciferase, the reporter gene for Renilla is a cDNA called Rluc that needs no post-translational modifications.
By coupling the operative regulatory elements to expression of a luciferase gene, typically by placing the regulatory element just upstream of a gene encoding luciferase, the cellular event can be detected readily by a luminescent signal. Thus, a cellular process or signal is available for study without directly assaying the cellular gene product. Typically, reporter genes are downstream of cloned response elements. When transfected into a cell, stronger promoters direct more luc mRNA synthesis and consequently, more protein is produced. The light intensity produced in the presence of luciferin allows the researcher to determine how strong a promoter is in the experimental system or how the promoter responds to a chemical treatment. View an animated introduction to reporter assays.
 Figure 1. Firefly and Renilla luciferase reactions with their respective substrates, beetle luciferin and coelenterazine, to yield light.
Figure 1. Firefly and Renilla luciferase reactions with their respective substrates, beetle luciferin and coelenterazine, to yield light. Optimized Luciferases
Native luciferases, such as firefly and Renilla, are not necessarily optimal as genetic reporters. We have enhanced the native proteins by improving their expression in mammalian systems, reducing the risk of anomalous expression, and in some cases, destabilizing them. To enhance mammalian expression, we have optimized codon usage for translation of the firefly and Renilla luciferase proteins. Additionally, inappropriate or unintended transcription regulatory sequences that may be used in mammalian cells have been minimized. The resulting recombinant luciferase reporters show much higher specific expression in mammalian systems.
The speed by which a genetic reporter can respond to changes in the transcriptional rate is correlated to the stability of the reporter within cells. Highly stable reporters accumulate to greater levels in cells, but their concentrations change slowly with changes in transcription. Conversely, lower stability yields less accumulation but a much faster rate of response. To provide reporters designed to meet different experimental needs, families of luciferase genes have been developed yielding different intracellular stabilities. Promega has developed destabilized luciferase reporters by genetically fusing a protein degradation sequence to the luciferase genes (3) . Due to their increased rate of degradation, these destabilized reporters respond faster and often display a greater magnitude of response to rapid transcriptional events. See our Rapid Response™ Reporters (Renilla and Firefly) for examples of these destabilized reporter vectors.
Dual-Reporter Gene Assays
Traditional reporter gene assays use a single reporter to monitor the transcriptional activity under investigation. A limitation of a single reporter assay is that under some experimental conditions, changes in luminescence can be because of factors other than the transcriptional control that is being studied. For example, when performing transient reporter expression, experimental discrepancies can occur due to well-to-well variability in transfection efficiency. Another issue arises when the experimental treatment reduces cell viability, resulting in nontranscriptional reduction in the luciferase signal.
The term “dual reporter” refers to the simultaneous expression and measurement of two individual reporter enzymes within a single system. Typically, the experimental reporter is correlated with the effect of specific experimental conditions, while the activity of the cotransfected control reporter provides an internal control that serves as the baseline response. Normalizing the activity of the experimental reporter to the activity of the internal control minimizes experimental variability caused by differences in cell viability or transfection efficiency. Other sources of variability, such as differences in pipetting volumes, cell lysis efficiency and assay efficiency, can be eliminated effectively as well. Thus, dual-reporter assays often allow more reliable interpretation of the experimental data. Firefly and Renilla luciferases, because of their distinct evolutionary origins, have dissimilar enzyme structures and substrate requirements. These differences make it possible to selectively discriminate between their respective bioluminescent reactions in a single tube without splitting the lysate.
Dual-reporter assays generally consist of: a firefly luciferase gene placed downstream from the response element of interest; a second reporter, Renilla luciferase, that is controlled by a housekeeping or viral promoter designed to be active under all conditions; and detection reagents that can sequentially measure luciferase activity. Two examples of such reagents that introduce the firefly substrate (luciferin), quench its signal and add the second substrate (coelentrazine) are the Dual-Luciferase® Reporter (DLR™) Assay and Dual-Glo® Assay (Figure 2). Cellular toxicity and variability in transfection efficiency can be controlled (normalized) by plotting the firefly induction as a ratio relative to the Renilla readout.
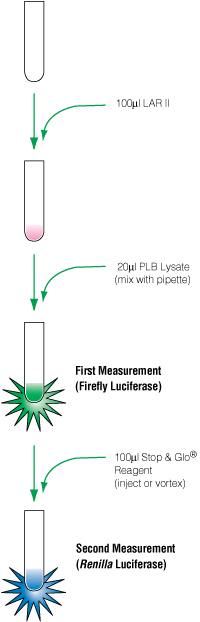 Figure 2. Flow chart of the DLR™ protocol.
Figure 2. Flow chart of the DLR™ protocol. Choosing a Reporter Vector
The starting point for any reporter assay is selecting the appropriate reporter vector, which is critical for the overall performance of the reporter assay. An array of luciferase reporter vectors exist because different biological systems work better with different vectors. Selecting a vector is based on your experimental conditions. Are you studying a response element like CRE, an enhancer you have just discovered, a newly defined promoter or a signaling pathway? Will your study require stable or transient transfection? Will you be measuring an endpoint or continuously monitoring the signal? Will a single luciferase assay give enough information depth, or is a dual-reporter assay more appropriate? Do you want a more responsive gene to study rapid transcriptional events, or do you want to generate the strongest luminescent signal possible? Understanding what you want to study and how you want to measure the results will help determine which vectors are most useful to you.
We provide a vast selection of firefly and Renilla luciferase vectors for all these reasons. In fact, the pGL4 Luciferase Reporter Vectors incorporate several advances in genetic reporter technology including codon optimization of the reporter gene to enhance expression in mammalian cells, inclusion of reporter genes that respond rapidly to transcriptional dynamics and incorporation of mammalian selection markers that facilitate stable cell line generation (4) (5) (6) . By selecting the right pGL4 Vector for your system and using any of our Luciferase Assay Systems, you can create an ultrasensitive assay that enables the accurate probing of the regulation and activity of a protein or a pathway inside the cell or in a cell lysate.
Applications of Bioluminescent Reporters
A common reporter gene setup will involve inserting a reporter gene downstream of a promoter/response element within a mammalian expression vector, which is then transfected into cultured cells. After treatment of the transfected cells, the effect of the biological stimulus can be measured by quantifying the amount of active luciferase that is expressed (Figure 3). Nevertheless, the broader aspect of gene expression entails much more than transcription alone, and reporter genes can be used to study these other cellular events (7) (8) . Figure 4 illustrates many of the applications to which our simple add-mix-measure genetic reporter assays can be applied including:
- Understanding promoter structure
- Activating/binding of surface receptors (e.g., GPCRs)
- Monitoring stem cell differentiation
- Analyzing transcription factor structure
- Observing viral infection
- Activating nuclear receptors
 Figure 3. Reporter assays are simple to design and easy to run.
Figure 3. Reporter assays are simple to design and easy to run.
 Figure 4. Promega has developed reporter gene assays for many key applications including gene transcription and regulation, receptor function and intracellular signaling, protein folding and metabolism, pathogen interactions with host cells, and RNAi suppression of gene function.
Figure 4. Promega has developed reporter gene assays for many key applications including gene transcription and regulation, receptor function and intracellular signaling, protein folding and metabolism, pathogen interactions with host cells, and RNAi suppression of gene function. Related Resources
Article References
- Fan, F. and Wood, K. (2007) Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 5, 127–36.
- Li, X. et al. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273, 34970–5.
- Almond, B.D. et al. (2004) Introducing the Rapid Response™ Reporter Vectors. Promega Notes 87, 18–22.
- Paguio, A. et al. (2005) pGL4 Vectors: A new generation of luciferase reporter vectors. Promega Notes 89, 7–10.
- Promega Corporation (2007) Deciphering the pGL4 Vector code. Promega Notes 96, 6–7.
- Wood, K. (1995) Marker proteins for gene expression. Curr. Opin. Biotechnol. 6, 50–8.
- Allard, S.T.M. and Kopish, K. (2008) Luciferase reporter assays: Powerful, adaptable tools for cell biology research. Cell Notes 21, 23–6.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Allard, S. T.M. Bioluminescent Reporter Genes. [Internet] October 2008. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/enotes/bioluminescent-reporter-genes/
American Medical Association, Manual of Style, 10th edition, 2007
Allard, S. T.M. Bioluminescent Reporter Genes. Promega Corporation Web site. https://www.promega.com/resources/pubhub/enotes/bioluminescent-reporter-genes/ Updated October 2008. Accessed Month Day, Year.
Products may be covered by pending or issued patents or may have certain limitations on use. Please visit our patent and trademark web page for more information.
 This animation introduces the concept of reporter gene assays.
This animation introduces the concept of reporter gene assays.