Using the Beta-Glo® Assay System to Determine Beta-galactosidase Activity in Yeast
Department of Biochemistry, University of Wisconsin, Madison, WI 53706
Publication Date: 2006
Abstract
The Beta-Glo® Assay System Cat.# E4720, E4740, E4780 was developed for rapid, sensitive and consistent quantitation of β-galactosidase in mammalian cells using a single-reagent addition. However, the experiments detailed in this article demonstrate that it can also be used to assay β-galactosidase expression in yeast two-hybrid and three-hybrid systems. The Beta-Glo® Reagent is used to rapidly lyse yeast cells and quantitate the β-galactosidase enzyme activity for detecting protein:protein or RNA:protein interactions. Because of the rapid lysis and quantitation and its multiwell format, the Beta-Glo® Assay System may also be adapted for high-throughput screening, enabling quicker screening for potential binding partners.
Introduction
The lacZ gene is often used as a reporter to monitor gene expression. Assays for the lacZ gene product, β-galactosidase, can be performed in numerous ways, including colorimetric assays on filter-grown yeast colonies and enzymatic assays of cleared yeast lysates. Here we describe using an enzymatic assay, the Beta-Glo® Assay System, in which the addition of a single reagent directly to yeast allows rapid quantitation of β-galactosidase. In particular, we use the Beta-Glo® Assay System with the yeast two- and three-hybrid systems.
The yeast, Saccharomyces cerevisiae, has become a vital tool for genetics, molecular biology and biochemistry. The yeast two- and three-hybrid systems are useful assays for identifying and analyzing protein:protein and RNA:protein interactions ( (1) ). In the yeast two-hybrid system, when the fusion protein, Protein X-LexA DNA binding domain, binds the fusion protein, Protein Y-transcription activation domain, lacZ , is transcribed (Figure 1, Panel A). This allows analysis of direct protein:protein interactions. In the yeast three-hybrid system, an RNA-binding protein (RBP) linked to a transcription activation domain (AD) is tested in combination with an RNA sequence (Figure 1, Panel B). An RNA:protein interaction results in activation of the HIS3 and lacZ reporter genes ( (1) (2) ). The system has been used to test candidate RNA-protein interactions, to isolate interaction-defective mutants, and to identify proteins that bind a given RNA sequence. Reverse yeast three-hybrid screens have also been used to identify binding sites for RNA-binding proteins ( (2) (3) (4) ).
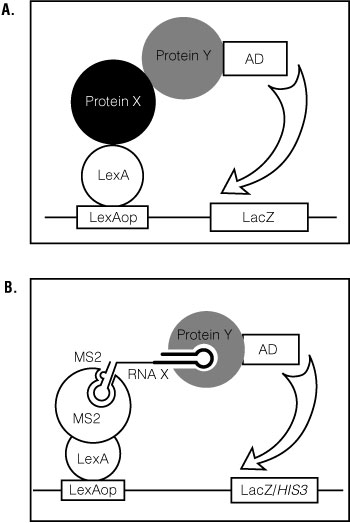 Figure 1. General schematic for the yeast two- and three-hybrid systems.
Figure 1. General schematic for the yeast two- and three-hybrid systems.
Panel A. In the yeast two-hybrid system, when the fusion protein, Protein X-LexA DNA binding domain, binds the fusion protein, Protein Y-transcription activation domain, lacZ, is transcribed. These interactions allow analysis of direct protein:protein interactions. Panel B. In the yeast three-hybrid system, an RNA-binding protein (RBP) linked to a transcription activation domain (AD) is tested in combination with an RNA sequence. An RNA:protein interaction results in activation of the HIS3 and lacZ reporter genes (1,2). The system has been used to test candidate RNA-protein interactions, to isolate interaction-defective mutants, and to identify proteins that bind a given RNA sequence. Reverse yeast three-hybrid screens have also been used to identify binding sites for RNA-binding proteins (2–4).
The “strength” of the interactions in the two- and three-hybrid systems is determined by assaying either β-galactosidase activity or by determining the level of resistance to 3-aminotriazole, which monitors HIS3 reporter activity. β-galactosidase can be assayed by measuring the conversion of a lactose analog to a chromogenic or luminescent product. This assay can be performed using either colonies permeabilized on a filter or in a cell lysate. The filter assay yields rapid but qualitative results; the liquid assay is more quantitative, but also more time consuming.
The Beta-Glo® Assay System allows rapid, sensitive and consistent quantitation of β-galactosidase. While the assay was originally developed and optimized for mammalian cells, we have adapted it for use in yeast. Because of the rapid lysis and quantitation, the Beta-Glo® Assay System has great potential for high-throughput screening of protein:protein and RNA-protein interactions.
Optimization of Beta-Glo® Assay System for Use in Yeast Three-Hybrid System
The Beta-Glo® Assay System was optimized in yeast using the three-hybrid system. An RBP (in this example, fem-3 binding factor (FBF) from C. elegans) was fused to a transcription activation domain, and various RNAs were fused to the MS2 stem loop sequence. First, we determined that Beta-Glo® Reagent lysed yeast, as required of the assay. Fifty microliters of yeast culture at an A600 of 0.1 was added to 50µl of Beta-Glo® Reagent, incubated for 30 minutes and plated on nonselective media. No colonies grew after 3 days at 30°C; many grew on the control plate [cells not treated with the reagent (not shown)].
Next, the optimal incubation time was determined. A reaction with 50µl of yeast at A600 of 0.1 was incubated with 50µl Beta-Glo® Reagent. After various incubation times, luminescence was measured (Figure 2, Panel A). The RNA sequences used in the assay bind the RBP with varying affinities. With all four RNAs, β-galactosidase activity plateaued at 60 minutes. For a strongly interacting RNA, RNA1 (Kd of 25nM), the signal decreased after 120 minutes. In all subsequent experiments, reactions were incubated for 60 minutes.
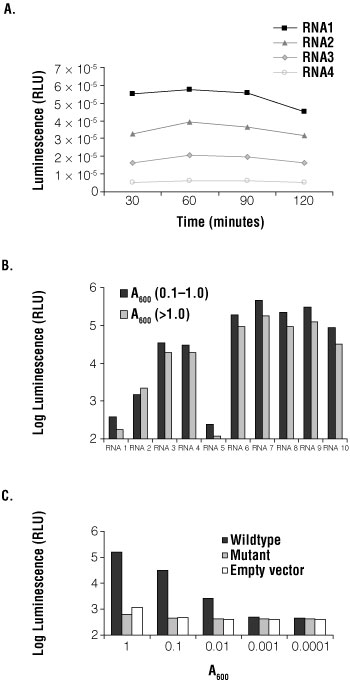 Figure 2. Optimization of the Beta-Glo Assay System in Saccharomyces cerevisiae.
Figure 2. Optimization of the Beta-Glo Assay System in Saccharomyces cerevisiae.
Panel A. Four RNA fusions and RBP-AD were transformed into yeast. Beta-Glo® Reagent was added to yeast grown to an A600 of 0.1 and incubated for 30, 60, 90 and 120 minutes. β-galactosidase activity was measured as luminescence and plotted as relative light units (RLU) versus time of incubation. Panel B. Ten RNA fusions and RBP-AD were transformed into yeast. Beta-Glo® Reagent was added to yeast grown to an A600 of 0.1 or to standing phase yeast (A600 >1.0) diluted to an A600 of 0.1 and incubated for 60 minutes. β-galactosidase activity was measured using a luminometer and represented as luminescence (RLU). The graph compared each RNA tested at both growth phases. RNAs do not correspond to those in Panel A. Panel C. A wild-type RNA fusion, mutant RNA fusion and empty vector were transformed into yeast with RBP-AD. The yeast were grown to an A600 of 1.0 and serially diluted to 0.0001. Beta-Glo® Reagent was incubated with yeast for 60 minutes, with an A600 range of 1.0, 0.1, 0.001, 0.001 and 0.0001. β-galactosidase activity was measured as luminescence and presented as RLU versus absorbance at 600nm.
To test the effect of growth phase on reporter detection, yeast were grown to low density (A600 of 0.1–1.0) and to high density (A600), and β-galactosidase activity was determined using the Beta-Glo® Assay System. Ten different RNAs, representing different affinity targets of the RBP, were tested at the two growth phases (Figure 2, Panel B). Yeast were grown to an A600 of 0.1 and to A600 of ~2.0. Yeast at high density were diluted to an A600 of 0.1. After a 60-minute incubation with Beta-Glo® Reagent, luminescence was measured. In all RNAs tested, the luminescence detected was greater with yeast at low density.
To determine the optimal number of yeast per assay, A600 versus signal-to-noise ratio was tested using three RNA constructs: wild-type RNA, a mutant RNA and empty vector. Yeast were grown to an A600 of 1.0 and serially diluted to 0.1, 0.01, 0.001, and 0.0001. Fifty microliters of diluted yeast was incubated with 50µl of Beta-Glo® Reagent for 60 minutes. The luminescence signal showed the assay was linear over a 10,000-fold range; however, an A600 of 1.0 had a larger signal-to-noise ratio than 0.1 (Figure 2, Panel C).
Optimized Protocol
Yeast cultures were grown in Selective Dropout media to low density (A600 of 0.1–1.0). Cultures were diluted to an A600 of 0.1. An equal volume of Beta-Glo® Reagent was then added, and the mixtures were incubated at room temperature for 60 minutes. The reactions were mixed and 10µl aliquoted into three Eppendorf tubes. The samples were then read in a luminometer (BD Monolight™ 2010 or Turner Biosystems 20/20) for one second.
Detecting β-galactosidase Activity in Yeast Three- and Two-Hybrid Systems with the Beta-Glo® Assay System
To test whether the Beta-Glo® Assay System used for the yeast three-hybrid system accurately reflected binding observed in other assays, the RBP-AD vector was transformed into yeast with empty vector, wild-type RNA and mutant RNA (Figure 3, Panel A). This RBP binds specifically to the wild-type sequence as determined by a β-galactosidase colony filter spot assay, HIS3 growth assay and electrophoretic mobility shift assay (EMSA; (2) (3) (4) (5) ). In the Beta-Glo® Assay System, the RBP bound specifically to its RNA target, but not the empty vector or mutant RNA. Thus the Beta-Glo® Assay System gave consistent results that corresponded well with other assays. Other combinations of RBPs and RNA targets yielded similar results (not shown).
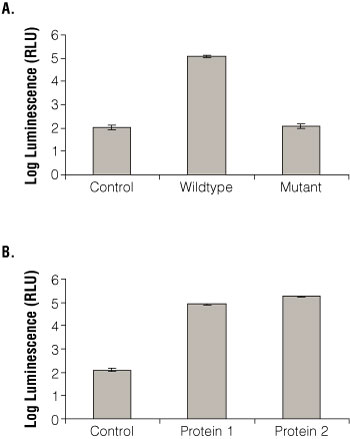 Figure 3. Beta-galactosidase activity determined using the Beta-Glo Assay System with the yeast three- and two-hybrid systems.
Figure 3. Beta-galactosidase activity determined using the Beta-Glo Assay System with the yeast three- and two-hybrid systems.
Panel A. Wild-type RNA fusion, mutant RNA fusion and empty RNA control vector were transformed into yeast with RBP-AD. Beta-Glo® Reagent was incubated with yeast grown to A600 of 0.1 for 60 minutes. β-galactosidase activity was measured with a luminometer and represented as luminescence (RLU). Panel B. Empty LexA DNA binding domain vector and two interacting fusion proteins were transformed into yeast with RBP-AD. Beta-Glo® Reagent was incubated with yeast grown to A600 of 0.1 for 60 minutes. As in Panel A, β-galactosidase activity was measured using a luminometer and represented as RLU.
The Beta-Glo® Assay System can also be used to measure β-galactosidase activity from the yeast two-hybrid system. Using the two-hybrid system of RBP-AD and the LexA DNA-binding domain vectors (empty vector and positive control proteins: Protein 1 and Protein 2), the Beta-Glo® Assay System was used to monitor protein:protein interaction in yeast (Figure 3, Panel B). The RBP appeared to interact with both proteins as expected from other independent data; it did not yield significant signal with the empty vector.
Conclusion
The Beta-Glo® Assay System can be used to assay β-galactosidase activity from yeast. Using the yeast three- and two-hybrid system, β-galactosidase activity was measured rapidly with high specificity and reproducibility. Optimal conditions for the Beta-Glo® Assay System were determined using an RBP and RNA sequences in the yeast three-hybrid system. The Beta-Glo® Assay System has already been used to detect interactions in the yeast three-hybrid system: RBP-RNA-directed tests ( (2) (3) (4) (6) (7) ) and RBP-RNA target screens ( (2) (3) (4) (6) ). This assay has several advantages over other β-galactosidase assays: it is easy, quick and sensitive with a large dynamic range (over four orders of magnitude; (8) ).
Related Products
Related Protocols
Related Resources
Article References
- Bernstein, D.S. et al. (2002) Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26, 123–41.
- Bernstein, D. et al. (2005) Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11, 447–58.
- Opperman, L. et al. (2005) A single spacer nucleotide determines the specificities of two mRNA regulatory proteins Nat. Struct. Mol. Biol. 12, 945–51.
- Crittenden, S.L. et al. (2002) A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660–3.
- Seay, D. et al. (2006) A three-hybrid screen identifies mRNAs controlled by a regulatory protein. RNA 12, 1594–600.
- MH Lee, B Hook, LB Lamont, M Wickens, and J Kimble (2006) LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans EMBO J 25, 88-96.
- Beta-Glo® Assay System Technical Manual, TM239, Promega Corporation
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Hook, B., Bernstein, D. and Wickens, M. Using the Beta-Glo Assay System to Determine Beta-galactosidase Activity in Yeast. [Internet] 2006. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/enotes/using-the-betaglo-assay-system-to-determine-beta-galactosidase-activity-in-yeast/
American Medical Association, Manual of Style, 10th edition, 2007
Hook, B., Bernstein, D. and Wickens, M. Using the Beta-Glo Assay System to Determine Beta-galactosidase Activity in Yeast. Promega Corporation Web site. https://www.promega.com/resources/pubhub/enotes/using-the-betaglo-assay-system-to-determine-beta-galactosidase-activity-in-yeast/ Updated 2006. Accessed Month Day, Year.
Products may be covered by pending or issued patents or may have certain limitations on use. Please visit our patent and trademark web page for more information.
 Learn about the chemistry of the bioluminescent reaction and its use in a broad range of assays.
Learn about the chemistry of the bioluminescent reaction and its use in a broad range of assays.