M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant: A Cost-Effective Alternative to SuperScript® II
Promega Corporation
Publication Date: October 2012
Abstract
Reverse transcriptases, also known as RNA-directed DNA polymerases, were first identified in retroviruses. In addition to the critical role these enzymes play in a variety of major human diseases (HIV, hepatitis and some cancers), reverse transcriptases are a fundamental component of the molecular biologists' toolbox. Reverse transcriptase activity is critical for many basic techniques including real-time and endpoint RT-PCR, labeled-probe generation for microarrays and cDNA cloning. The ideal reverse transcriptase is robust, with high activity under a variety of conditions, and converts all primed RNA within a sample to cDNA, regardless of the RNA abundance, length or secondary structure. In this report, we show that M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant from Promega has similar thermal stability properties, produces long cDNA transcripts and performs similarly in RT-PCR to SuperScript® II, making it a good alternative reverse transcriptase.
Introduction
Moloney Murine Leukemia Virus Reverse Transcriptase, RNase H Minus, Point Mutant [M-MLV (H–), Point Mutant], is an RNA-dependent DNA polymerase that can be used in cDNA synthesis with long RNA templates (>5kb). RNase H activity is germane to this application because RNase H activity can degrade template when incubation times are long, and long incubation times are often necessary when making long cDNAs. Here we compare the Promega M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant (Cat.# M3681) to SuperScript® II (Life Technologies) for the ability to produce long cDNA transcripts. We also compare the performance of these two reverse transcriptases in RT-PCR, RT-qPCR and label incorporation experiments.
Materials
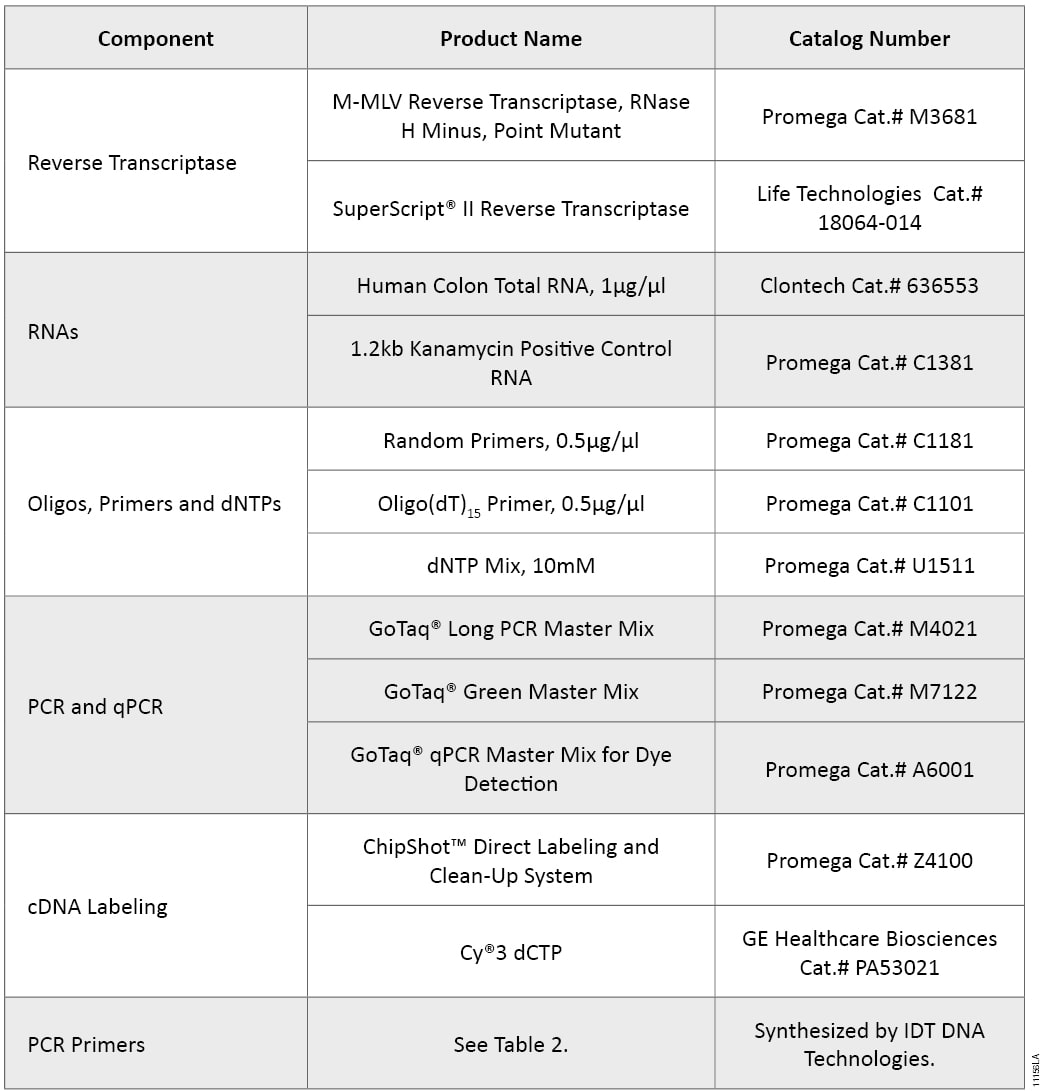 Table 1. Products Used for Reverse Transcription, PCR/qPCR and DNA Labeling.
Table 1. Products Used for Reverse Transcription, PCR/qPCR and DNA Labeling.
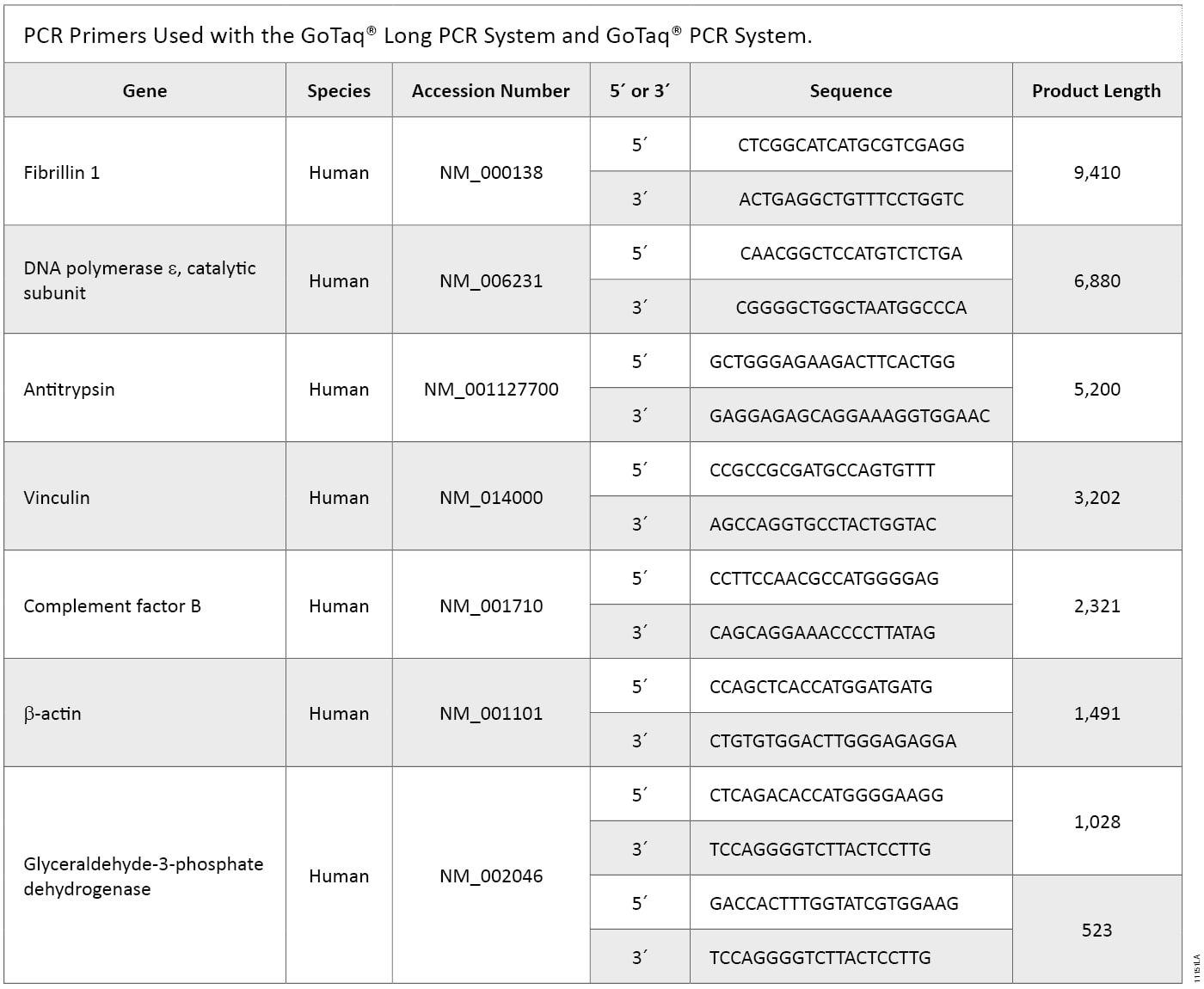 Table 2. Primers Used in GoTaq® Long PCR and GoTaq® PCR Systems.
Table 2. Primers Used in GoTaq® Long PCR and GoTaq® PCR Systems. Methods
Reverse Transcription Reactions: Reactions were performed as described in the manufacturers’ instructions for each product. All reactions were incubated at 42°C for 50 minutes unless otherwise stated. GoTaq® Long PCR amplifications were prepared as described in Table 3, using 1µl of the reverse transcription reactions as template. See Technical Manual #TM359 for details.
Table 3. GoTaq® Long PCR Master Mix Thermal Cycling Guidelines.
|
Step |
Temperature |
Time |
| Initial Denaturation |
92°C |
2 minutes |
| Denaturation |
92°C |
30 seconds |
| Annealing/Extension |
65°C |
15 minutes |
| Repeat Denaturation/Annealing/ Extension for total of 35 cycles |
— |
— |
| Final Extension |
72°C |
10 minutes |
| Soak |
4°C |
— |
GoTaq® Green Master Mix (Cat.# M7122) PCR amplifications were prepared as described in Table 4. The 0.5kb GAPDH primers were added to the master mix along with 2µl of the reverse transcription reactions. See the GoTaq® Green Master Mix Product Information Sheet, #9PIM712, for details.
Table 4. GoTaq® Green Master Mix Thermal Cycling Guidelines.
|
Step |
Temperature |
Time |
| Initial Denaturation |
95°C |
2 minutes |
| Denaturation |
95°C |
30 seconds |
| Annealing |
52°C |
30 seconds |
| Extension |
72°C |
1 minute |
| Repeat Denaturation/Annealing/ Extension for total of 40 cycles |
— |
— |
| Final Extension |
72°C |
5 minutes |
| Soak |
4°C |
— |
GoTaq® qPCR amplifications were prepared as described in Table 5, using 5µl of the reverse transcription reactions as template, and primers designed to amplify the kanamycin control template. See Technical Manual #TM318 for details. A Bio-Rad CFX real-time thermal cycler was used with the conditions listed in Table 5.
Table 5. GoTaq® qPCR Thermal Cycling Guidelines.
|
Step |
Temperature |
Time |
| Hot-Start Inactivation |
95°C |
2 minutes |
| Denaturation |
95°C |
15 seconds |
| Annealing/Extension |
60°C |
1 minute |
| Repeat Denaturation/Annealing/ Extension for total of 45 cycles |
— |
— |
| Dissociation |
60-95°C |
— |
Agarose gel analysis was performed using 1% agarose gels (Agarose, LE, Analytical Grade, Promega Cat.# V3121) containing 0.4% ethidium bromide solution (Ethidium Bromide Solution, Molecular Grade, Promega Cat.# H5041). Gels were run at 100V. A Bio-Rad XR imager was used to analyze gels.
Results
Thermal Stability and Synthesis of Large Transcripts
M-MLV (H–), Point Mutant, and SuperScript® II reverse transcriptases are reported to have maximal activity at 42°C, with reduced activity at 50°C. We performed reverse transcription reactions at 37°C, 42°C and 50°C, followed by GoTaq® Long PCR with five primer sets (each a different transcript) that amplified products 9.4kb, 6.9kb, 5.2kb, 3.2kb and 1.5kb in length. The resulting PCR products were analyzed using agarose gel electrophoresis (Figure 1). Reverse transcription reactions at 42°C for both enzymes resulted in products for all five primer sets. Increasing the temperature to 50°C reduced the amount of product accumulated. Both enzymes performed similarly under these conditions.
 Figure 1. Thermal stability and synthesis of large transcripts.
Figure 1. Thermal stability and synthesis of large transcripts. cDNAs were synthesized from 1µg of total human RNA using M-MLV (H–), Point Mutant, (Panel A) or SuperScript® II (Panel B) reverse transcriptase at 37°C, 42°C and 50°C. GoTaq® Long PCR amplifications were performed on each transcription reaction using five primer sets, amplifying products from 9.4kb to 1.5kb. Ten microliters of the 9.4, 6.9, and 5.2kb PCR products and 1µl of the 3.2 and 1.5kb PCR products were separated on a 1% agarose gel. Lane M. BenchTop 1kb DNA Ladder (Cat.# G7541).
Synthesis of Transcripts of Varying Lengths
Reverse transcription reactions were performed at the reported optimal 42°C using 1µg of total human RNA for both M-MLV (H–), Point Mutant, and SuperScript® II reverse transcriptases. We used the GoTaq® Long PCR System and eight primer sets to amplify 1µl of the cDNA produced from the reverse transcription reactions. The amplification reactions were analyzed on a 1% agarose gel (Figure 2). Both systems were able to amplify products from all eight primer sets including a large 9.4kb transcript, Fibrillin. Only a small amount of the shortest product, 0.5kb GAPDH, was synthesized because the GoTaq® Long PCR System is optimized for longer transcripts. M-MLV (H–), Point Mutant, and SuperScript® II reverse transcriptases performed similarly and were able to synthesize transcripts of varying sizes, including very long transcripts.
 Figure 2. Synthesis of transcripts of various lengths.
Figure 2. Synthesis of transcripts of various lengths. cDNAs were synthesized from 1µg of total human RNA using M-MLV (H–), Point Mutant, (Panel A) or SuperScript®II (Panel B) reverse transcriptase at each enzyme’s optimal temperature. GoTaq® Long PCR amplification was performed on each transcription reaction using eight primer sets, amplifying products from 9.4kb to 0.5kb in size. Ten microliters of the 9.4, 6.9, and 5.2kb PCR products and 1µl of the 3.2, 1.5, 1.0 and 0.5kb PCR products were separated on a 1% agarose gel. Lane M. BenchTop 1kb DNA Ladder (Cat.# G7541).
High-Quality cDNA from Very Small Amounts of Total RNA
Decreasing amounts of Human Total Colon RNA (100ng–100fg) were used in first-strand cDNA synthesis reactions with either M-MLV (H–), Point Mutant, or SuperScript® II reverse transcriptase. Following cDNA synthesis, amplification reactions were performed using 5µl of the reverse transcription reactions and primers for a 0.5kb fragment of GAPDH. The PCR products were visualized using agarose gel electrophoresis (Figure 3). Both enzymes were able to synthesize cDNA from even the lowest amount of template tested, 100fg of total RNA.
 Figure 3. Generating high-quality cDNA from very small amounts of total RNA.
Figure 3. Generating high-quality cDNA from very small amounts of total RNA. Total human RNA was serially diluted (100ng–100fg) and used as template in reverse transcription reactions with M-MLV (H–), Point Mutant,( Panel A) or SuperScript® II (Panel B) reverse transcriptase. GoTaq® PCR amplification was performed on the transcripts using GAPDH-specific primers (0.5kb product size). Twenty microliters of the PCR products were separated on a 1% agarose gel. Lane M. BenchTop 1kb DNA Ladder (Cat.# G7541).
Sensitivity in qPCR
We spiked decreasing amounts (1 x 1010 to 10 molecules) of a single transcript RNA into 1ng of human total RNA and performed reverse transcription with 50 units of M-MLV (H–), Point Mutant, and SuperScript® II reverse transcriptases. An aliquot of the reverse transcription reactions (5µl) was amplified using GoTaq® qPCR Master Mix (Figure 4). Transcripts resulting from both enzymes produced amplification products when starting with as little as 1 x 103 molecules. M-MLV (H–) Point Mutant was similar to SuperScript® II in producing cDNAs from low concentrations of RNA and amplifying in a qPCR amplification.
 Figure 4. Sensitivity in a qPCR amplification.
Figure 4. Sensitivity in a qPCR amplification. Kanamycin control RNA was serially diluted tenfold (1 × 1010 to 1 × 101), spiked into 1ng of total human RNA and used as template in reverse transcription reactions with M-MLV (H–), Point Mutant, (Panel A) or SuperScript®II (Panel B) reverse transcriptase. Quantitative PCR amplification was performed on 5µl of the GoTaq® qPCR System with kanamycin control primers. Cq values are plotted versus molecules of RNA in the reverse transcription reactions. Cq values could not be determined at amounts less than 1 × 103 molecules. N=3.
Label Incorporation
We tested the ability of each reverse transcriptase to incorporate a fluorescently labeled nucleotide into the synthesized cDNA, an approach commonly used in microarray experiments. The ChipShot™ Direct Labeling and Clean-Up System (Promega Cat.# Z4000) provided a positive control for labeling as well as a method for isolating the cDNA from non-incorporated dye-labeled nucleotides. The reverse transcriptase provided with the system was used as a positive control, and a reaction containing no reverse transcriptase (–RT) was used was a negative control. Each cleaned-up, labeled cDNA reaction was analyzed on a NanoDrop®-1000 using absorbance detection at A550 and A260. The amount (ng) of synthesized cDNA and the frequency of incorporation were calculated and plotted (Figure 5). M-MLV (H–) Point Mutant Reverse Transcriptase produced 32% more cDNA than SuperScript® II and had 26% higher frequency of dye incorporation.
 Figure 5. Label incorporation.
Figure 5. Label incorporation. Reverse transcription reactions were performed as described in the ChipShot™ Direct Labeling and Clean-Up System Technical Manual, #TM286, using the positive control transcriptase in the kit and M-MLV (H–), Point Mutant, or SuperScript® II reverse transcriptase. A no-reverse transcriptase reaction (–RT) was also performed as a negative control. Reactions were cleaned-up using the ChipShot™ Direct Labeling and Clean-Up System (Cat.# Z4100). The amount of cDNA generated and dye incorporated were determined using A260 and A550 values. N=2.
Conclusions
M-MLV (RNase H Minus) Point Mutant Reverse Transcriptase displayed similar properties and abilities compared to SuperScript® II reverse transcriptase, making M-MLV (H–) Point Mutant Reverse Transcriptase an equivalent, cost-effective alternative. M-MLV (H–), Point Mutant,was stable at 42°C and able to produce cDNA from small (0.5kb) to large (9.4kb) RNA transcripts. M-MLV (H–), Point Mutant, was highly sensitive, producing cDNAs from low concentrations of RNA as analyzed using end-point PCR and qPCR. DNA dye-based labeling was efficiently performed using M-MLV (H–), Point Mutant, Reverse Transcriptase, producing more cDNA and a higher frequency of dye incorporation as compared to SuperScript® II reverse transcriptase.
Related Products
| Product |
Size |
Catalog Number |
| GoTaq® Long PCR Master Mix |
100 reactions |
|
| GoTaq® Green Master Mix |
100 reactions |
|
| Agarose, LE, Analytical Grade |
100g |
|
| Ethidium Bromide Solution, Molecular Grade |
10mg |
|
| BenchTop 1kb DNA Ladder |
600µl |
Related Products
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Hook, B., Bratz, M. and Schagat, T. M-MLV Reverse Transcriptase RNase H Minus Point Mutant: A Cost-Effective Alternative to SuperScript® II. [Internet] October 2012. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/mmlv-reverse-transcriptase-rnase-h-minus-point-mutant-a-cost-effective-alternative/
American Medical Association, Manual of Style, 10th edition, 2007
Hook, B., Bratz, M. and Schagat, T. M-MLV Reverse Transcriptase RNase H Minus Point Mutant: A Cost-Effective Alternative to SuperScript® II. Promega Corporation Web site. https://www.promega.com/resources/pubhub/mmlv-reverse-transcriptase-rnase-h-minus-point-mutant-a-cost-effective-alternative/ Updated October 2012. Accessed Month Day, Year.