Purifying miRNA from Blood and Cell Culture Using the Maxwell RSC miRNA Tissue Kit
Promega Corporation
Publication Date: Feb 2016; tpub_169
Abstract
The challenges associated with purifying miRNA vary depending upon the source material. For example, cells or tissue samples often have miRNAs in relatively abundant numbers, whereas circulating miRNAs in extracellular fluid, such as plasma or serum, may be relatively scarce. Blood samples may pose additional problems, due to high levels of endogenous RNases. There are multiple methods for purifying miRNA. These methods are generally similar to those for total RNA, but RNA purification is followed by an additional step during which the miRNA fraction is isolated and enriched by size-selection or selective precipitation. Most commercial methods are based on organic extraction of total RNA and purification of miRNA on silica columns. The Maxwell® RSC miRNA Tissue Kit overcomes the limitations of organic extraction, producing purified total RNA including miRNA suitable for downstream analysis by a variety of techniques. Here we evaluate the Maxwell® RSC miRNA Tissue Kit performance in purifying miRNA from whole blood and cell culture samples in terms of the yield, integrity and purity of the RNA obtained.
Materials and Methods
miRNA from Blood
For miRNA purification from whole blood, 2.5ml of fresh whole blood held in an EDTA collection tube was added to a 15ml tube. Cell Lysis Solution (7.5ml; Part# A793A) was added, and the tube was inverted 5–6 times to mix. The lysate was incubated for 10 minutes at room temperature. Twice during the incubation, the tube was inverted to mix. The sample was centrifuged at 3,000 x g for 10 minutes. The supernatant was removed and discarded without disturbing the visible white pellet. The sample was briefly spun to collect residual liquid at the bottom of the tube, and the remaining supernatant was discarded.
Chilled 1-Thioglycerol/Homogenization Solution (200µl) was added to the pellet, and the tube was vortexed to resuspend the pellet. Next, 200µl of Lysis Buffer and 25µl of Proteinase K solution were added. Samples were vortexed and incubated at room temperature for 10 minutes. Finally, the samples were loaded into well #1 of the Maxwell® cartridge, and 10µl of DNaseI was added to well #4. The RNA was purified with the Maxwell® RSC Instrument using the microRNA Tissue Kit v4.99 method and eluted in 60µl of Nuclease-Free Water.
miRNA from Cell Culture
Cultured HEK293, Jurkat and NIH3T3 cells (106 cells) were pelleted, the media was removed, and the cell pellets were stored at –80°C until they could be processed. Chilled 1-Thioglycerol/Homogenization Solution (200µl) was added to the pellet, and the tubes were vortexed to resuspend the pellets. Next, 200µl of Lysis Buffer and 15µl of Proteinase K solution were added. Samples were vortexed and incubated at room temperature for 10 minutes. Finally, the samples were loaded into well #1 of the Maxwell® cartridge, and 10µl of DNase I was added to well #4. The RNA was purified with the Maxwell® RSC Instrument using the microRNA Tissue Kit v4.99 method and eluted in 60µl of Nuclease-Free Water.
All RNA samples were stored at –80°C until they could be analyzed.
Results
RNA Concentration, Yield and Purity
Concentration, yield and purity of the purified RNA was determined using the NanoDrop®-1000 and the QuantiFluor® RNA System on the Quantus™ Fluorometer. RNA integrity numbers (RIN) were determined using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer.
The results for concentration, yield and purity of the RNA samples are shown in Figures 1–3. Both blood and cultured cells produced RNA in high concentrations and yields (Figures 1 and 2). RNA purity for most samples averaged >1.90 with the exception of Jurkat cells, which averaged ~1.80 and had greater variability among replicates (Figure 3). Finally, RIN values (Figure 1) were lower than expected for HEK293 cells (7.8) and blood (6.8), compared with NIH3T3 cells (9.3) and Jurkat cells (9.0).
 Figure 1. RNA concentration and integrity (RIN).
Figure 1. RNA concentration and integrity (RIN). RNA concentration was determined using the NanoDrop®-1000, and the QuantiFluor® RNA System on the Quantus™ Fluorometer. RIN values were determined using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer. N = 4. Standard deviations are shown.
 Figure 2. RNA yields. Yield values were determined using the NanoDrop®-1000 and the QuantiFluor® RNA System on the Quantus™ Fluorometer. N = 4. Standard deviations are shown.
Figure 2. RNA yields. Yield values were determined using the NanoDrop®-1000 and the QuantiFluor® RNA System on the Quantus™ Fluorometer. N = 4. Standard deviations are shown. Figure 3. RNA purity.
Figure 3. RNA purity. Values were determined using the NanoDrop®-1000. N = 4. Standard deviations are shown.
miRNA Yields
To determine the yield of miRNA, RT-qPCR reactions were prepared using the following miRNA-specific oligos: miR-21 (Life Technologies 000397), miR-125b (Life Technologies 000449), miR-141 (Life Technologies 000463) and let7e (Life Technologies 0002406). Oligos consisting of the miRNA sequences were diluted to 1 × 107, 1 × 106, 1 × 105, 1 × 104 and 1 × 103 copies/µl in Nuclease-Free Water to serve as a standard to determine miRNA copy number recovered per 2.5ml of whole blood or 1 million cells.
RNA samples were diluted to 2ng/µl in Nuclease-Free Water. TaqMan® reverse transcription reactions were set up. Ten microliters of the reverse transcription master mix was added to 5µl of the RNA template or 5µl of the miRNA oligo standards. For the no-target control (NTC) reaction, 5µl of Nuclease-Free Water was added. Samples were run on the Bio-Rad CFX96™ Real-Time PCR System.
Following reverse transcription, qPCR was performed using TaqMan® miRNA-specific primers for each miRNA target. The RT reaction (1.3µl in duplicate) was added to 18.7µl of the GoTaq® Probe qPCR Master Mix and the samples were run on the Bio-Rad CFX96™ Real-Time PCR System.
The results are shown in Figure 4. microRNA was successfully purified from blood and all cell types, although the yields of microRNAs varied between sample types and which miRNA was targeted.
 Figure 4. RT-qPCR using miRNA-specific primers.
Figure 4. RT-qPCR using miRNA-specific primers. RT-qPCR was performed on purified RNA using the TaqMan® miRNA-specific primers. microRNA yield is given in copies per 2.5ml of whole blood or 1 million cells. N = 4. Standard deviations are shown.
Purified mRNA Performance in Real-Time RT-PCR
To test the performance of the purified messenger RNA in amplification reactions, real-time PCR was performed using 10ng of RNA (diluted in Nuclease-Free Water) in the GoTaq® 1-Step RT-qPCR System using mouse beta-2-microglobulin primers (5’ CTATATCCTGGCTCACACTGAAT 3’; 5’ CTTGATCACATGTCTCGATCCCA 3’) and human beta-2-microglobulin primers (5’ TGCTGTCTCCATGTTTGATGTATCT 3’; 5’ TCTCTGCTCCCCACCTCTAAGT 3’) or the GoTaq® Probe 1-Step RT-qPCR System with mouse lamin A TaqMan® primers (Life Technologies Mm00497783_m1 Lmna) or human HPRT1 TaqMan® primers (Life Technologies Hs03929098_m1).
GoTaq® 1-Step RT-qPCR System
Fifteen microliters of the master mix was added to 5µl of the RNA template. For the no-template control reaction, 5µl of Nuclease-Free Water was added. Samples were run on the Bio-Rad CFX96™ Real-Time PCR System.
GoTaq® Probe 1-Step RT-qPCR System
Fifteen microliters of the master mix was added to 5µl of the RNA template. For the no-template control reaction, 5µl of Nuclease-Free Water was added. Samples were run on the Bio-Rad CFX96™ Real-Time PCR System.
The results are shown in Figure 5. mRNAs were successfully purified from blood and all cell types. The relative amount of mRNAs varied between sample types.
 Figure 5. RT-qPCR results from purified mRNA.
Figure 5. RT-qPCR results from purified mRNA. RT-qPCR was performed using 10ng of purified RNA using mouse and human beta-2-microglobulin primers with the GoTaq® 1-Step RT-qPCR System and mouse lamin A or human HPRT1 TaqMan® primers with the GoTaq® Probe 1-Step RT-qPCR System. Hs = human. Mm = mouse. N = 4. Standard deviations are shown.
Spike Test for Inhibition
We tested for the presence of inhibitors in the purified RNA using the Solaris™ RNA Spike Control Kit (GE Healthcare #K-002200-C1-100). Purified RNA or Nuclease-Free Water (No Inhibition Control; NIC) was added to a GoTaq® Probe 1-Step RT-qPCR System containing the RNA spike control template and primers.
To calculate inhibition, we subtracted the average Cq values of the NIC samples from the average Cq values of the samples with the purified RNA. The results are shown in Table 1. No inhibition is evident in the RNA purified from the blood or any of the cell types.
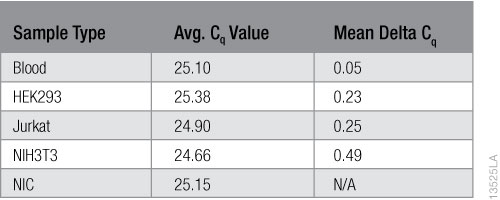 Table 1. RNA inhibition Results using the Solaris™ RNA Spike Control Kit.
Table 1. RNA inhibition Results using the Solaris™ RNA Spike Control Kit. Conclusions
We evaluated the Maxwell® RSC miRNA Tissue Kit performance for purifying miRNA from whole blood and cell culture samples. The kit purified high concentrations and yields of RNA from blood and cells. The RNA purity was generally high with A260/A230 averages >1.90. The only exception we observed was for Jurkat cells, which averaged ~1.80 for A260/A230 ratios and had greater variability among replicates. We used the TaqMan® MicroRNA Assay and four different miRNA-specific oligos to verify that miRNA was successfully purified from all the sample types, although the yield (copy number) of miRNA varied by sample type and the specific miRNA targeted. We successfully purified mRNA from blood and all cell types. The relative amount of mRNA purified and the RIN values varied between sample types as well as cell types. The RNA integrity numbers for 3T3 cells and Jurkat cells fell between 9 and 10, and showed the least variability. The RIN values for HEK293 cells fell between 7 and 8, and those for blood were the lowest, averaging below 7. None of the purified RNA samples showed any evidence of amplification inhibition using the Solaris™ RNA Spike Control Kit and the GoTaq® Probe 1-Step RT-qPCR System.
Related Products
Related Protocols
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
D. Wieczorek and E. Lepinski Purifying miRNA from Blood and Cell Culture Using the Maxwell® RSC miRNA Tissue Kit. [Internet] Feb 2016; tpub_169. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/purifying-mirna-from-blood-and-cell-culture-with-maxwell-rscmi-rna-tissue-kit/
American Medical Association, Manual of Style, 10th edition, 2007
D. Wieczorek and E. Lepinski Purifying miRNA from Blood and Cell Culture Using the Maxwell® RSC miRNA Tissue Kit. Promega Corporation Web site. https://www.promega.com/resources/pubhub/purifying-mirna-from-blood-and-cell-culture-with-maxwell-rscmi-rna-tissue-kit/ Updated Feb 2016; tpub_169. Accessed Month Day, Year.
GoTaq, Maxwell and QuantiFluor are registered trademarks of Promega Corporation. Quantus is a trademark of Promega Corporation.
CFX96 is a trademark of Bio-Rad Laboratories, Inc. NanoDrop is a registered trademark of Thermo Fisher Scientific, Inc. Solaris is a trademark of Thermo Fisher Scientific, Inc. TaqMan is a registered trademark of Roche Molecular Systems, Inc.